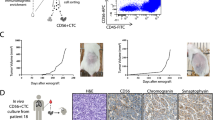
Abstract
In most patients with small-cell lung cancer (SCLC)—a metastatic, aggressive disease—the condition is initially chemosensitive but then relapses with acquired chemoresistance. In a minority of patients, however, relapse occurs within 3 months of initial treatment; in these cases, disease is defined as chemorefractory. The molecular mechanisms that differentiate chemosensitive from chemorefractory disease are currently unknown. To identify genetic features that distinguish chemosensitive from chemorefractory disease, we examined copy-number aberrations (CNAs) in circulating tumor cells (CTCs) from pretreatment SCLC blood samples. After analysis of 88 CTCs isolated from 13 patients (training set), we generated a CNA-based classifier that we validated in 18 additional patients (testing set, 112 CTC samples) and in six SCLC patient-derived CTC explant tumors1. The classifier correctly assigned 83.3% of the cases as chemorefractory or chemosensitive. Furthermore, a significant difference was observed in progression-free survival (PFS) (Kaplan–Meier P value = 0.0166) between patients designated as chemorefractory or chemosensitive by using the baseline CNA classifier. Notably, CTC CNA profiles obtained at relapse from five patients with initially chemosensitive disease did not switch to a chemorefractory CNA profile, which suggests that the genetic basis for initial chemoresistance differs from that underlying acquired chemoresistance.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Hodgkinson, C.L. et al. Tumorigenicity and genetic profiling of circulating tumor cells in small-cell lung cancer. Nat. Med. 20, 897–903 (2014).
Rudin, C.M. et al. Comprehensive genomic analysis identifies SOX2 as a frequently amplified gene in small-cell lung cancer. Nat. Genet. 44, 1111–1116 (2012).
Peifer, M. et al. Integrative genome analyses identify key somatic driver mutations of small-cell lung cancer. Nat. Genet. 44, 1104–1110 (2012).
George, J. et al. Comprehensive genomic profiles of small cell lung cancer. Nature 524, 47–53 (2015).
Lawrence, M.S. et al. Mutational heterogeneity in cancer and the search for new cancer-associated genes. Nature 499, 214–218 (2013).
Semenova, E.A., Nagel, R. & Berns, A. Origins, genetic landscape, and emerging therapies of small cell lung cancer. Genes Dev. 29, 1447–1462 (2015).
Dowlati, A. et al. Clinical correlation of extensive-stage small-cell lung cancer genomics. Ann. Oncol. 27, 642–647 (2016).
Hou, J.M. et al. Evaluation of circulating tumor cells and serological cell death biomarkers in small cell lung cancer patients undergoing chemotherapy. Am. J. Pathol. 175, 808–816 (2009).
Tanaka, F. et al. Circulating tumor cell as a diagnostic marker in primary lung cancer. Clin. Cancer Res. 15, 6980–6986 (2009).
Hou, J.M. et al. Clinical significance and molecular characteristics of circulating tumor cells and circulating tumor microemboli in patients with small-cell lung cancer. J. Clin. Oncol. 30, 525–532 (2012).
Chan, B.A. & Coward, J.I.G. Chemotherapy advances in small-cell lung cancer. J. Thorac. Dis. 5 (Suppl. 5), S565–S578 (2013).
Polzer, B. et al. Molecular profiling of single circulating tumor cells with diagnostic intention. EMBO Mol. Med. 6, 1371–1386 (2014).
Smyth, G.K. Linear models and empirical Bayes methods for assessing differential expression in microarray experiments. Stat. Appl. Genet. Mol. Biol. 3, 1544–6115 1027.
von Pawel, J. et al. Topotecan versus cyclophosphamide, doxorubicin, and vincristine for the treatment of recurrent small-cell lung cancer. J. Clin. Oncol. 17, 658–667 (1999).
O'Brien, M.E. et al. Phase III trial comparing supportive care alone with supportive care with oral topotecan in patients with relapsed small-cell lung cancer. J. Clin. Oncol. 24, 5441–5447 (2006).
Hagmann, R., Hess, V., Zippelius, A. & Rothschild, S.I. Second-line therapy of small-cell lung cancer: topotecan compared to a combination treatment with adriamycin, cyclophosphamide and vincristine (ACO)—a single center experience. J. Cancer 6, 1148–1154 (2015).
van Meerbeeck, J.P., Fennell, D.A. & De Ruysscher, D.K. Small-cell lung cancer. Lancet 378, 1741–1755 (2011).
Lally, B.E., Urbanic, J.J., Blackstock, A.W., Miller, A.A. & Perry, M.C. Small cell lung cancer: have we made any progress over the last 25 years? Oncologist 12, 1096–1104 (2007).
Fruh, M. et al. & on behalf of the ESMO Guidelines Working Group. Small-cell lung cancer (SCLC): ESMO clinical practice guidelines for diagnosis, treatment and follow-up. Ann. Oncol. 24 (Suppl. 6), Vi99–Vi105 (2013).
White, S.C. et al. Randomized phase II study of cyclophosphamide, doxorubicin, and vincristine compared with single-agent carboplatin in patients with poor prognosis small cell lung carcinoma. Cancer 92, 601–608 (2001).
Boeva, V. et al. Control-free calling of copy number alterations in deep-sequencing data using GC-content normalization. Bioinformatics 27, 268–269 (2011).
Yates, T., Okoniewski, M.J. & Miller, C.J. X:Map: annotation and visualization of genome structure for Affymetrix exon array analysis. Nucleic Acids Res. 36, D780–D786 (2008).
R Core Team. R: A language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria. Available at: http://www.R-project.org/ (2014).
Kuhn, M. Building predictive models in R using the caret package. J. Stat. Softw. 28, 1–26 (2008).
Racine, J.C. RSTUDIO: A platform-independent IDE for R and sweave. J. Appl. Econ. 27, 167–172 (2012).
Acknowledgements
L.C. was funded via a Clinical Pharmacology Fellowship educational grant from CRUK and AstraZeneca (C147/A12328). We would like to acknowledge the help of the CRUK Molecular Biology Core Facilities and members of the CRUK Manchester Institute Clinical and Experimental Pharmacology Group for their support of this study. We also thank the patients and healthy volunteers who provided their blood samples. This work was supported by core funding to CRUK Manchester Institute (C5759/A12328), and via Manchester CRUK Centre Award (A12197) and a research grant from Menarini Biomarkers Singapore PTE Ltd. (funding for all assigned to C.D.)
Author information
Authors and Affiliations
Contributions
G.B., F.B., A.H. and C.D. supervised and devised the study, interpreted data and co-wrote the manuscript. L.C., D.G.R. and B.M. interpreted data, co-wrote the manuscript and planned and ran the WGA and NGS experiments. C.S., H.S.L., F.F.-G., Y.L. and C.M. performed the bioinformatics analysis. D.J.B. and J.A. ran the DEPArray isolations and optimized the WGA pipeline. C.J.M. and C.L.H. generated the CDX models. K.M. managed the CellSearch CTC analysis. L.C., M.C., L.P. and F.B. oversaw the acquisition of ethical permission and patient consent and the collection of blood samples. All authors read and approved the final manuscript.
Corresponding authors
Ethics declarations
Competing interests
A. Hughes was previously employed by AstraZeneca and holds shares in AstraZeneca. A. Hughes is currently employed by the University of Manchester.
Supplementary information
Supplementary Text, Figures and Tables
Supplementary Figures 1–6 and Supplementary Tables 1–5 (PDF 4531 kb)
Supplementary Table 6
Excel spread sheet of 2281 loci identified in 16 profiles of CTC CNA predictive signature. (XLSX 30 kb)
Rights and permissions
About this article
Cite this article
Carter, L., Rothwell, D., Mesquita, B. et al. Molecular analysis of circulating tumor cells identifies distinct copy-number profiles in patients with chemosensitive and chemorefractory small-cell lung cancer. Nat Med 23, 114–119 (2017). https://doi.org/10.1038/nm.4239
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nm.4239
This article is cited by
-
Single-cell low-pass whole genome sequencing accurately detects circulating tumor cells for liquid biopsy-based multi-cancer diagnosis
npj Precision Oncology (2024)
-
Resistance to BRAF inhibition explored through single circulating tumour cell molecular profiling in BRAF-mutant non-small-cell lung cancer
British Journal of Cancer (2024)
-
Genetically-engineered mouse models of small cell lung cancer: the next generation
Oncogene (2024)
-
Research progress on the multi-omics and survival status of circulating tumor cells
Clinical and Experimental Medicine (2024)
-
The functional and clinical roles of liquid biopsy in patient-derived models
Journal of Hematology & Oncology (2023)