Abstract
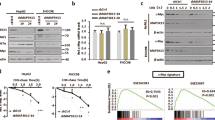
MYC oncoproteins are involved in the genesis and maintenance of the majority of human tumors but are considered undruggable. By using a direct in vivo shRNA screen, we show that liver cancer cells that have mutations in the gene encoding the tumor suppressor protein p53 (Trp53 in mice and TP53 in humans) and that are driven by the oncoprotein NRAS become addicted to MYC stabilization via a mechanism mediated by aurora kinase A (AURKA). This MYC stabilization enables the tumor cells to overcome a latent G2/M cell cycle arrest that is mediated by AURKA and the tumor suppressor protein p19ARF. MYC directly binds to AURKA, and inhibition of this protein–protein interaction by conformation-changing AURKA inhibitors results in subsequent MYC degradation and cell death. These conformation-changing AURKA inhibitors, with one of them currently being tested in early clinical trials, suppressed tumor growth and prolonged survival in mice bearing Trp53-deficient, NRAS-driven MYC-expressing hepatocellular carcinomas (HCCs). TP53-mutated human HCCs revealed increased AURKA expression and a positive correlation between AURKA and MYC expression. In xenograft models, mice bearing TP53-mutated or TP53-deleted human HCCs were hypersensitive to treatment with conformation-changing AURKA inhibitors, thus suggesting a therapeutic strategy for this subgroup of human HCCs.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
Accession codes
Primary accessions
BioProject
Referenced accessions
Gene Expression Omnibus
Protein Data Bank
References
Eilers, M. & Eisenman, R.N. Myc's broad reach. Genes Dev. 22, 2755–2766 (2008).
Meyer, N. & Penn, L.Z. Reflecting on 25 years with MYC. Nat. Rev. Cancer 8, 976–990 (2008).
Soucek, L. et al. Modeling Myc inhibition as a cancer therapy. Nature 455, 679–683 (2008).
Shachaf, C.M. et al. MYC inactivation uncovers pluripotent differentiation and tumor dormancy in hepatocellular cancer. Nature 431, 1112–1117 (2004).
Felsher, D.W. & Bishop, J.M. Reversible tumorigenesis by MYC in hematopoietic lineages. Mol. Cell 4, 199–207 (1999).
Podsypanina, K., Politi, K., Beverly, L.J. & Varmus, H.E. Oncogene cooperation in tumor maintenance and tumor recurrence in mouse mammary tumors induced by Myc and mutant Kras. Proc. Natl. Acad. Sci. USA 105, 5242–5247 (2008).
Zuber, J. et al. RNAi screen identifies Brd4 as a therapeutic target in acute myeloid leukemia. Nature 478, 524–528 (2011).
Delmore, J.E. et al. BET bromodomain inhibition as a therapeutic strategy to target c-Myc. Cell 146, 904–917 (2011).
Otto, T. et al. Stabilization of NMYC is a critical function of Aurora A in human neuroblastoma. Cancer Cell 15, 67–78 (2009).
Brockmann, M. et al. Small-molecule inhibitors of Aurora A induce proteasomal degradation of N-MYC in childhood neuroblastoma. Cancer Cell 24, 75–89 (2013).
Gustafson, W.C. et al. Drugging MYCN through an allosteric transition in aurora kinase A. Cancer Cell 26, 414–427 (2014).
Beltran, H. et al. Molecular characterization of neuroendocrine prostate cancer and identification of new drug targets. Cancer Discov. 1, 487–495 (2011).
Mosquera, J.M. et al. Concurrent AURKA and MYCN gene amplifications are harbingers of lethal-treatment-related neuroendocrine prostate cancer. Neoplasia 15, 1–10 (2013).
Bruix, J., Gores, G.J. & Mazzaferro, V. Hepatocellular carcinoma: clinical frontiers and perspectives. Gut. 63, 844–855 (2014).
Lord, R., Suddle, A. & Ross, P.J. Emerging strategies in the treatment of advanced hepatocellular carcinoma: the role of targeted therapies. Int. J. Clin. Pract. 65, 182–188 (2011).
Welsch, C., Jesudian, A., Zeuzem, S. & Jacobson, I. New direct-acting antiviral agents for the treatment of hepatitis C virus infection and perspectives. Gut 61 (suppl. 1), i36–i46 (2012).
Calle, E.E. & Kaaks, R. Overweight, obesity and cancer: epidemiological evidence and proposed mechanisms. Nat. Rev. Cancer 4, 579–591 (2004).
White, D.L., Kanwal, F. & El-Serag, H.B. Association between non-alcoholic fatty liver disease and risk for hepatocellular cancer, based on systematic review. Clin. Gastroenterol. Hepatol. 10, 1342–1359.e2 (2012).
Liu, J. et al. Alterations of TP53 are associated with a poor outcome for patients with hepatocellular carcinoma: evidence from a systematic review and meta-analysis. Eur. J. Cancer 48, 2328–2338 (2012).
Chan, K.L., Guan, X.Y. & Ng, I.O. High-throughput tissue microarray analysis of c-Myc activation in chronic liver diseases and hepatocellular carcinoma. Hum. Pathol. 35, 1324–1331 (2004).
Meek, D.W. Tumor suppression by p53: a role for the DNA damage response? Nat. Rev. Cancer 9, 714–723 (2009).
Sherr, C.J. Tumor surveillance via the ARF–p53 pathway. Genes Dev. 12, 2984–2991 (1998).
Tannapfel, A. et al. INK4A–ARF alterations and p53 mutations in hepatocellular carcinomas. Oncogene 20, 7104–7109 (2001).
Sherr, C.J. Divorcing ARF and p53: an unsettled case. Nat. Rev. Cancer 6, 663–673 (2006).
Carlson, C.M., Frandsen, J.L., Kirchhof, N., McIvor, R.S. & Largaespada, D.A. Somatic integration of an oncogene-harboring Sleeping Beauty transposon models liver tumor development in the mouse. Proc. Natl. Acad. Sci. USA 102, 17059–17064 (2005).
Kang, T.W. et al. Senescence surveillance of premalignant hepatocytes limits liver cancer development. Nature 479, 547–551 (2011).
Wuestefeld, T. et al. A direct in vivo RNAi screen identifies MKK4 as a key regulator of liver regeneration. Cell 153, 389–401 (2013).
Tward, A.D. et al. Genomic progression in mouse models for liver tumors. Cold Spring Harb. Symp. Quant. Biol. 70, 217–224 (2005).
Rudalska, R. et al. In vivo RNAi screening identifies a mechanism of sorafenib resistance in liver cancer. Nat. Med. 20, 1138–1146 (2014).
Calvisi, D.F. et al. Ubiquitous activation of RAS and JAK–STAT pathways in human HCC. Gastroenterology 130, 1117–1128 (2006).
Schmidt, C.M., McKillop, I.H., Cahill, P.A. & Sitzmann, J.V. Increased MAPK expression and activity in primary human hepatocellular carcinoma. Biochem. Biophys. Res. Commun. 236, 54–58 (1997).
Ito, Y. et al. Activation of mitogen-activated protein kinases–extracellular signal-regulated kinases in human hepatocellular carcinoma. Hepatology 27, 951–958 (1998).
Crane, R., Gadea, B., Littlepage, L., Wu, H. & Ruderman, J.V. Aurora A, meiosis and mitosis. Biol. Cell 96, 215–229 (2004).
Katayama, H., Brinkley, W.R. & Sen, S. The aurora kinases: role in cell transformation and tumorigenesis. Cancer Metastasis Rev. 22, 451–464 (2003).
Katsha, A., Belkhiri, A., Goff, L. & El-Rifai, W. Aurora kinase A in gastrointestinal cancers: time to target. Mol. Cancer 14, 106 (2015).
Lu, L.Y. et al. Aurora A is essential for early embryonic development and tumor suppression. J. Biol. Chem. 283, 31785–31790 (2008).
Kimura, M.T. et al. Two functional coding single-nucleotide polymorphisms in STK15 (aurora A) coordinately increase esophageal cancer risk. Cancer Res. 65, 3548–3554 (2005).
Khwaja, A., Rodriguez-Viciana, P., Wennström, S., Warne, P.H. & Downward, J. Matrix adhesion and Ras transformation both activate a phosphoinositide 3-OH kinase and protein kinase B (AKT) cellular survival pathway. EMBO J. 16, 2783–2793 (1997).
Klein, A., Flügel, D. & Kietzmann, T. Transcriptional regulation of serine/threonine kinase 15 (STK15) expression by hypoxia and HIF-1. Mol. Biol. Cell 19, 3667–3675 (2008).
Farazi, P.A. & DePinho, R.A. Hepatocellular carcinoma pathogenesis: from genes to environment. Nat. Rev. Cancer 6, 674–687 (2006).
Pérez Tamayo, R. Is cirrhosis of the liver experimentally produced by CCl4 an adequate model of human cirrhosis? Hepatology 3, 112–120 (1983).
Taub, R. Liver regeneration 4: transcriptional control of liver regeneration. FASEB J. 10, 413–427 (1996).
White, P., Brestelli, J.E., Kaestner, K.H. & Greenbaum, L.E. Identification of transcriptional networks during liver regeneration. J. Biol. Chem. 280, 3715–3722 (2005).
Yuan, H. et al. Overcoming CML acquired resistance by specific inhibition of Aurora A kinase in the KCL-22 cell model. Carcinogenesis 33, 285–293 (2012).
Carpinelli, P. et al. PHA-739358, a potent inhibitor of aurora kinases with a selective target inhibition profile relevant to cancer. Mol. Cancer Ther. 6, 3158–3168 (2007).
Aliagas-Martin, I. et al. A class of 2,4-bisanilinopyrimidine Aurora A inhibitors with unusually high selectivity against Aurora B. J. Med. Chem. 52, 3300–3307 (2009).
Fancelli, D. et al. 1,4,5,6-tetrahydropyrrolo[3,4-c]pyrazoles: identification of a potent aurora kinase inhibitor with a favorable antitumor kinase inhibition profile. J. Med. Chem. 49, 7247–7251 (2006).
Falchook, G. et al. Investigational Aurora A kinase inhibitor alisertib (MLN8237) as an enteric-coated tablet formulation in nonhematologic malignancies: phase 1 dose-escalation study. Invest. New Drugs 32, 1181–1187 (2014).
Amati, B. Myc degradation: dancing with ubiquitin ligases. Proc. Natl. Acad. Sci. USA 101, 8843–8844 (2004).
Eilers, M., Schirm, S. & Bishop, J.M. The MYC protein activates transcription of the α-prothymosin gene. EMBO J. 10, 133–141 (1991).
Watson, J.D., Oster, S.K., Shago, M., Khosravi, F. & Penn, L.Z. Identifying genes regulated in a Myc-dependent manner. J. Biol. Chem. 277, 36921–36930 (2002).
Lu, L. et al. Aurora kinase A mediates c-Myc's oncogenic effects in hepatocellular carcinoma. Mol. Carcinog. 54, 1467–1479 (2015).
Zender, L. et al. An oncogenomics-based in vivo RNAi screen identifies tumor suppressors in liver cancer. Cell 135, 852–864 (2008).
Burgess, D.J. et al. Topoisomerase levels determine chemotherapy response in vitro and in vivo. Proc. Natl. Acad. Sci. USA 105, 9053–9058 (2008).
Braumüller, H. et al. T helper 1 cell cytokines drive cancer into senescence. Nature 494, 361–365 (2013).
Huesken, D. et al. Design of a genome-wide siRNA library using an artificial neural network. Nat. Biotechnol. 23, 995–1001 (2005).
Vert, J.P., Foveau, N., Lajaunie, C. & Vandenbrouck, Y. An accurate and interpretable model for siRNA efficacy prediction. BMC Bioinformatics 7, 520 (2006).
Zender, L. et al. Caspase-8 small interfering RNA prevents acute liver failure in mice. Proc. Natl. Acad. Sci. USA 100, 7797–7802 (2003).
Krizhanovsky, V. et al. Senescence of activated stellate cells limits liver fibrosis. Cell 134, 657–667 (2008).
Diefenbacher, M.E. et al. Usp28 counteracts Fbw7 in intestinal homeostasis and cancer. Cancer Res. 75, 1181–1186 (2015).
Acknowledgements
We thank K. Schulze-Osthoff, S.W. Lowe, W. Albrecht, K. Breuhahn, R. Geffers, S. Nahnsen, M. Gehringer, E. Rist, C. Fellmeth, P. Katiyar, S. Fillinger, M. Jarek, M. Scharfe, M. Diefenbacher and the members of the Zender laboratory for fruitful discussions, technical assistance or reagents. We thank the Center for Scientific Computing (Espoo, Finland) for software and hardware support. This work was supported by the German Research Foundation (DFG; grants FOR2314 (L.Z., D.D. and M.E.) and SFB685 (L.Z.)), the Gottfried Wilhelm Leibniz Program (L.Z.), the European Research Council (projects 'CholangioConcept' (L.Z.), 'Heptromic' (L.Z.) and 'Auromyc' (M.E.)), the German Ministry for Education and Research (BMBF) (eMed (Multiscale HCC), the German Universities Excellence Initiative (third funding line: 'future concept') (L.Z.), the German Center for Translational Cancer Research (DKTK) (L.Z.) and the German–Israeli Cooperation in Cancer Research (DKFZ–MOST) (L.Z.).
Author information
Authors and Affiliations
Contributions
L.Z. and D.D. designed the study; D.D., R.R., G.C., T.-W.K., T.W., A.H., T.Y., L.H. and P.B. conducted the research; T.L. performed the histopathological analyses; J.-C.N., S.I., T.L. and J.Z.-R. collected and analyzed the human HCC samples; T.P. and A.P. performed the molecular modeling; N.P.M., S.L. and M.E. contributed to research design; and L.Z. and D.D wrote the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–13 (PDF 9476 kb)
Supplementary Table 1
mRNA Expression data (XLS 7070 kb)
Supplementary Table 2
Genes included in the shRNA library (XLS 36 kb)
Rights and permissions
About this article
Cite this article
Dauch, D., Rudalska, R., Cossa, G. et al. A MYC–aurora kinase A protein complex represents an actionable drug target in p53-altered liver cancer. Nat Med 22, 744–753 (2016). https://doi.org/10.1038/nm.4107
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nm.4107
This article is cited by
-
Deficiency of BAP1 inhibits neuroblastoma tumorigenesis through destabilization of MYCN
Cell Death & Disease (2023)
-
Evolving therapeutic landscape of advanced hepatocellular carcinoma
Nature Reviews Gastroenterology & Hepatology (2023)
-
Circlehunter: a tool to identify extrachromosomal circular DNA from ATAC-Seq data
Oncogenesis (2023)
-
MYC in liver cancer: mechanisms and targeted therapy opportunities
Oncogene (2023)
-
Potent molecular-targeted therapies for gastro-entero-pancreatic neuroendocrine carcinoma
Cancer and Metastasis Reviews (2023)