Abstract
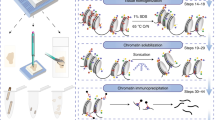
Extensive cross-linking introduced during routine tissue fixation of clinical pathology specimens severely hampers chromatin immunoprecipitation followed by next-generation sequencing (ChIP-seq) analysis from archived tissue samples. This limits the ability to study the epigenomes of valuable, clinically annotated tissue resources. Here we describe fixed-tissue chromatin immunoprecipitation sequencing (FiT-seq), a method that enables reliable extraction of soluble chromatin from formalin-fixed paraffin-embedded (FFPE) tissue samples for accurate detection of histone marks. We demonstrate that FiT-seq data from FFPE specimens are concordant with ChIP-seq data from fresh-frozen samples of the same tumors. By using multiple histone marks, we generate chromatin-state maps and identify cis-regulatory elements in clinical samples from various tumor types that can readily allow us to distinguish between cancers by the tissue of origin. Tumor-specific enhancers and superenhancers that are elucidated by FiT-seq analysis correlate with known oncogenic drivers in different tissues and can assist in the understanding of how chromatin states affect gene regulation.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
Accession codes
References
Garraway, L.A. & Lander, E.S. Lessons from the cancer genome. Cell 153, 17–37 (2013).
Fanelli, M. et al. Pathology tissue–chromatin immunoprecipitation, coupled with high-throughput sequencing, allows the epigenetic profiling of patient samples. Proc. Natl. Acad. Sci. USA 107, 21535–21540 (2010).
Barski, A. et al. High-resolution profiling of histone methylations in the human genome. Cell 129, 823–837 (2007).
Shi, S.R., Imam, S.A., Young, L., Cote, R.J. & Taylor, C.R. Antigen retrieval immunohistochemistry under the influence of pH using monoclonal antibodies. J. Histochem. Cytochem. 43, 193–201 (1995).
Shi, S.R., Liu, C., Perez, J. & Taylor, C.R. Protein-embedding technique: a potential approach to standardization of immunohistochemistry for formalin-fixed, paraffin-embedded tissue sections. J. Histochem. Cytochem. 53, 1167–1170 (2005).
Yamashita, S. Heat-induced antigen retrieval: mechanisms and application to histochemistry. Prog. Histochem. Cytochem. 41, 141–200 (2007).
Shin, H., Liu, T., Duan, X., Zhang, Y. & Liu, X.S. Computational methodology for ChIP-seq analysis. Quant. Biol. 1, 54–70 (2013).
Anders, S., Pyl, P.T. & Huber, W. HTSeq—a Python framework to work with high-throughput sequencing data. Bioinformatics 31, 166–169 (2015).
Anders, S. & Huber, W. Differential expression analysis for sequence-count data. Genome Biol. 11, R106 (2010).
Lee, T.I. et al. Control of developmental regulators by Polycomb in human embryonic stem cells. Cell 125, 301–313 (2006).
Ben-Porath, I. et al. An embryonic stem-cell-like gene expression signature in poorly differentiated aggressive human tumors. Nat. Genet. 40, 499–507 (2008).
Hansen, K.D. et al. Increased methylation variation in epigenetic domains across cancer types. Nat. Genet. 43, 768–775 (2011).
Balasubramanian, D. et al. H3K4me3 inversely correlates with DNA methylation at a large class of non-CpG-island-containing start sites. Genome Med. 4, 47 (2012).
Cancer Genome Atlas Network. Comprehensive molecular characterization of human colon and rectal cancer. Nature 487, 330–337 (2012).
Carroll, J.S. et al. Genome-wide analysis of estrogen-receptor-binding sites. Nat. Genet. 38, 1289–1297 (2006).
Ernst, J. & Kellis, M. ChromHMM: automating chromatin-state discovery and characterization. Nat. Methods 9, 215–216 (2012).
Kundaje, A. et al. Integrative analysis of 111 reference human epigenomes. Nature 518, 317–330 (2015).
Verzi, M.P. et al. TCF4 and CDX2, major transcription factors for intestinal function, converge on the same cis-regulatory regions. Proc. Natl. Acad. Sci. USA 107, 15157–15162 (2010).
Verzi, M.P. et al. Differentiation-specific histone modifications reveal dynamic chromatin interactions and partners for the intestinal transcription factor CDX2. Dev. Cell 19, 713–726 (2010).
Hatzis, P. et al. Genome-wide pattern of TCF7L2 (TCF4) chromatin occupancy in colorectal cancer cells. Mol. Cell. Biol. 28, 2732–2744 (2008).
van Bragt, M.P., Hu, X., Xie, Y. & Li, Z. RUNX1, a transcription factor mutated in breast cancer, controls the fate of ER-positive mammary luminal cells. eLife 3, e03881 (2014).
Santagata, S., Ligon, K.L. & Hornick, J.L. Embryonic stem cell transcription factor signatures in the diagnosis of primary and metastatic germ cell tumors. Am. J. Surg. Pathol. 31, 836–845 (2007).
Zhang, X., Zhang, J., Wang, T., Esteban, M.A. & Pei, D. Esrrb activates Oct4 transcription and sustains self-renewal and pluripotency in embryonic stem cells. J. Biol. Chem. 283, 35825–35833 (2008).
Heng, J.C. et al. The nuclear receptor Nr5a2 can replace Oct4 in the reprogramming of murine somatic cells to pluripotent cells. Cell Stem Cell 6, 167–174 (2010).
Hnisz, D. et al. Superenhancers in the control of cell identity and disease. Cell 155, 934–947 (2013).
Zentner, G.E. & Scacheri, P.C. The chromatin fingerprint of gene enhancer elements. J. Biol. Chem. 287, 30888–30896 (2012).
Ong, C.T. & Corces, V.G. Enhancer function: new insights into the regulation of tissue-specific gene expression. Nat. Rev. Genet. 12, 283–293 (2011).
Kouros-Mehr, H., Slorach, E.M., Sternlicht, M.D. & Werb, Z. GATA-3 maintains the differentiation of the luminal cell fate in the mammary gland. Cell 127, 1041–1055 (2006).
Jiang, J. et al. A core Klf circuitry regulates self-renewal of embryonic stem cells. Nat. Cell Biol. 10, 353–360 (2008).
Cancer Genome Atlas Research Network. Comprehensive molecular characterization of urothelial bladder carcinoma. Nature 507, 315–322 (2014).
Robinson, J.T. et al. Integrative genomics viewer. Nat. Biotechnol. 29, 24–26 (2011).
Ramírez, F., Dündar, F., Diehl, S., Grüning, B.A. & Manke, T. deepTools: a flexible platform for exploring deep-sequencing data. Nucleic Acids Res. 42, W187–W191 (2014).
Liu, T. et al. Cistrome: an integrative platform for transcriptional regulation studies. Genome Biol. 12, R83 (2011).
McLean, C.Y. et al. GREAT improves functional interpretation of cis-regulatory regions. Nat. Biotechnol. 28, 495–501 (2010).
Heinz, S. et al. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol. Cell 38, 576–589 (2010).
Subramanian, A. et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. USA 102, 15545–15550 (2005).
Trapnell, C. et al. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat. Protoc. 7, 562–578 (2012).
Acknowledgements
This work was supported by the DFCI–Novartis Drug Discovery Program (R.A.S.), the US NIH grant P50CA127003 (R.A.S.), the Susan F. Smith Center for Women's Cancers (M. Brown), grants from the Susan G. Komen and the Breast Cancer Research foundations (M. Brown), the Medical Oncology Translational Grant Program from the Dana-Farber Cancer Institute (C.J.S.), a grant from Instituto de Salud Carlos III the Spanish Economy and Competitiveness Ministry (grant PI13-01818; P.C.), the ConSEPOC–Comunidad de Madrid (grant S2010/BMD-2542; P.C.), and fellowships from the Asociación Española Contra el Cáncer (Programa de Formación Avanzada en Oncología 2010; P.C.) and the Fundación Caja Madrid (P.C.). We thank E. Díaz and M. de Miguel for technical assistance.
Author information
Authors and Affiliations
Contributions
P.C., R.A.S., M. Brown and H.W.L. conceived and designed the study; L.L., P.C., H.W.L. and N.K.O'N. performed computational and statistical analyses; M.D., P.R., M. Bowden and C.W.Z. acquired data; E.B. and M.M. reviewed CRC pathology; J.F., J.M.-R., H.G., V.M., S.M., J.B., D.G.-O., C.J.S., M.H. and A.R. supervised collection of tumor samples; X.S.L., M. Brown, H.W.L. and R.A.S. provided overall supervision; and P.C., R.A.S. and H.W.L. drafted the manuscript, with input from all of the authors.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–4 (PDF 3763 kb)
Supplementary Table 1
Sample characteristics, study design, and ChIP-seq library statistics. (XLSX 28 kb)
Supplementary Table 2
Comparison of PAT-ChIP-seq and FiT-seq performance on FFPE samples. (XLSX 25 kb)
Supplementary Table 3
Genes differentially H3K4me2 marked at promoters between normal and tumoral colorectal samples. (XLSX 1866 kb)
Supplementary Table 4
Gene Set Enrichment Analysis of the differentially H3K4me2 marked promoters between normal and tumoral colorectal samples for both the FF and FFPE cohorts. (XLSX 1682 kb)
Supplementary Table 5
Motif analysis at the differentially marked enhancers between normal and tumoral colorectal samples. (XLSX 55 kb)
Supplementary Table 6
Locations of the most discriminant enhancers across-tumortypes as determined by ANOVA. (XLSX 340 kb)
Supplementary Table 7
Motif analysis at the most discriminant enhancers acrosstumor-types. (XLSX 97 kb)
Supplementary Table 8
List of super-enhancers called for the series of tumors included in the cross tumor study in Figure 4. (XLSX 9396 kb)
Rights and permissions
About this article
Cite this article
Cejas, P., Li, L., O'Neill, N. et al. Chromatin immunoprecipitation from fixed clinical tissues reveals tumor-specific enhancer profiles. Nat Med 22, 685–691 (2016). https://doi.org/10.1038/nm.4085
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nm.4085
This article is cited by
-
Epigenomic analysis of formalin-fixed paraffin-embedded samples by CUT&Tag
Nature Communications (2023)
-
FiTAc-seq: fixed-tissue ChIP-seq for H3K27ac profiling and super-enhancer analysis of FFPE tissues
Nature Protocols (2020)
-
CDK4/6 inhibition reprograms the breast cancer enhancer landscape by stimulating AP-1 transcriptional activity
Nature Cancer (2020)
-
Enhanced and controlled chromatin extraction from FFPE tissues and the application to ChIP-seq
BMC Genomics (2019)
-
Enhancer signatures stratify and predict outcomes of non-functional pancreatic neuroendocrine tumors
Nature Medicine (2019)