Abstract
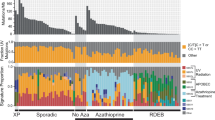
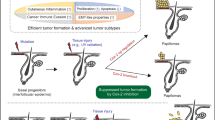
Mouse models of cancers are routinely used to study cancer biology. However, it remains unclear whether carcinogenesis in mice is driven by the same spectrum of genomic alterations found in humans. Here we conducted a comprehensive genomic analysis of 9,10-dimethyl-1,2-benzanthracene (DMBA)-induced skin cancer, the most commonly used skin cancer model, which appears as benign papillomas that progress into squamous cell carcinomas (SCCs). We also studied genetically induced SCCs that expressed G12D mutant Kras (Kras G12D) but were deficient for p53. Using whole-exome sequencing, we uncovered a characteristic mutational signature of DMBA-induced SCCs. We found that the vast majority of DMBA-induced SCCs presented recurrent mutations in Hras, Kras or Rras2 and mutations in several additional putative oncogenes and tumor-suppressor genes. Similar genes were recurrently mutated in mouse and human SCCs that were from different organs or had been exposed to different carcinogens. Invasive SCCs, but not papillomas, presented substantial chromosomal aberrations, especially in DMBA-induced and genetically induced Trp53-mutated SCCs. Metastasis occurred through sequential spreading, with relatively few additional genetic events. This study provides a framework for future functional cancer genomic studies in mice.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
Accession codes
Change history
06 August 2015
In the published article, the format was listed as Article, but this is a Resource. The error has been corrected in the HTML and PDF versions of the article.
References
Luch, A. Nature and nurture —lessons from chemical carcinogenesis. Nat. Rev. Cancer 5, 113–125 (2005).
Poirier, M.C. Chemical-induced DNA damage and human cancer risk. Nat. Rev. Cancer 4, 630–637 (2004).
Abel, E.L., Angel, J.M., Kiguchi, K. & DiGiovanni, J. Multi-stage chemical carcinogenesis in mouse skin: fundamentals and applications. Nat. Protoc. 4, 1350–1362 (2009).
Balmain, A., Ramsden, M., Bowden, G.T. & Smith, J. Activation of the mouse cellular Harvey-ras gene in chemically induced benign skin papillomas. Nature 307, 658–660 (1984).
Quintanilla, M., Brown, K., Ramsden, M. & Balmain, A. Carcinogen-specific mutation and amplification of Ha-ras during mouse skin carcinogenesis. Nature 322, 78–80 (1986).
Bizub, D., Wood, A.W. & Skalka, A.M. Mutagenesis of the Ha-ras oncogene in mouse skin tumors induced by polycyclic aromatic hydrocarbons. Proc. Natl. Acad. Sci. USA 83, 6048–6052 (1986).
Ruggeri, B. et al. Alterations of the p53 tumor suppressor gene during mouse skin tumor progression. Cancer Res. 51, 6615–6621 (1991).
Kemp, C.J., Donehower, L.A., Bradley, A. & Balmain, A. Reduction of p53 gene dosage does not increase initiation or promotion but enhances malignant progression of chemically induced skin tumors. Cell 74, 813–822 (1993).
Aldaz, C.M., Trono, D., Larcher, F., Slaga, T.J. & Conti, C.J. Sequential trisomization of chromosomes 6 and 7 in mouse skin premalignant lesions. Mol. Carcinog. 2, 22–26 (1989).
Quigley, D.A. et al. Network analysis of skin tumor progression identifies a rewired genetic architecture affecting inflammation and tumor susceptibility. Genome Biol. 12, R5 (2011).
Lapouge, G. et al. Identifying the cellular origin of squamous skin tumors. Proc. Natl. Acad. Sci. USA 108, 7431–7436 (2011).
White, A.C. et al. Defining the origins of Ras/p53-mediated squamous cell carcinoma. Proc. Natl. Acad. Sci. USA 108, 7425–7430 (2011).
Beck, B. et al. A vascular niche and a VEGF-Nrp1 loop regulate the initiation and stemness of skin tumours. Nature 478, 399–403 (2011).
Alexandrov, L.B. et al. Signatures of mutational processes in human cancer. Nature 500, 415–421 (2013).
Dipple, A., Pigott, M., Moschel, R.C. & Costantino, N. Evidence that binding of 7,12-dimethylbenz(a)anthracene to DNA in mouse embryo cell cultures results in extensive substitution of both adenine and guanine residues. Cancer Res. 43, 4132–4135 (1983).
Cai, Y., Patel, D.J., Broyde, S. & Geacintov, N.E. Base sequence context effects on nucleotide excision repair. J. Nucleic Acids 2010, 174252 (2010).
Westcott, P.M. et al. The mutational landscapes of genetic and chemical models of Kras-driven lung cancer. Nature 517, 489–492 (2015).
Bos, J.L. ras oncogenes in human cancer: a review. Cancer Res. 49, 4682–4689 (1989).
Saez, R., Chan, A.M., Miki, T. & Aaronson, S.A. Oncogenic activation of human R-ras by point mutations analogous to those of prototype H-ras oncogenes. Oncogene 9, 2977–2982 (1994).
Schramek, D. et al. Direct in vivo RNAi screen unveils myosin IIa as a tumor suppressor of squamous cell carcinomas. Science 343, 309–313 (2014).
Chu, W.K. & Hickson, I.D. RecQ helicases: multifunctional genome caretakers. Nat. Rev. Cancer 9, 644–654 (2009).
Nicolas, M. et al. Notch1 functions as a tumor suppressor in mouse skin. Nat. Genet. 33, 416–421 (2003).
Laczmanska, I. et al. Copy number alterations of chromosomal regions enclosing protein tyrosine phosphatase receptor-like genes in colorectal cancer. Pathol. Res. Pract. 210, 893–896 (2014).
Provost, E. et al. The tumor suppressor rpl36 restrains KRAS(G12V)-induced pancreatic cancer. Zebrafish 11, 551–559 (2014).
The Cancer Genome Atlas Network. Comprehensive genomic characterization of head and neck squamous cell carcinomas. Nature 517, 576–582 (2015).
Cohen-Dvashi, H. et al. Navigator-3, a modulator of cell migration, may act as a suppressor of breast cancer progression. EMBO Mol. Med. 7, 299–314 (2015).
Dickinson, R.E. et al. Epigenetic inactivation of SLIT3 and SLIT1 genes in human cancers. Br. J. Cancer 91, 2071–2078 (2004).
Hase, K. et al. AP-1B-mediated protein sorting regulates polarity and proliferation of intestinal epithelial cells in mice. Gastroenterology 145, 625–635 (2013).
Yu, J. et al. Kibra functions as a tumor suppressor protein that regulates Hippo signaling in conjunction with Merlin and Expanded. Dev. Cell 18, 288–299 (2010).
Pogue-Geile, K.L. et al. Palladin mutation causes familial pancreatic cancer and suggests a new cancer mechanism. PLoS Med. 3, e516 (2006).
Lee, C.S. et al. Recurrent point mutations in the kinetochore gene KNSTRN in cutaneous squamous cell carcinoma. Nat. Genet. 46, 1060–1062 (2014).
South, A.P. et al. NOTCH1 mutations occur early during cutaneous squamous cell carcinogenesis. J. Invest. Dermatol. 134, 2630–2638 (2014).
Pickering, C.R. et al. Mutational landscape of aggressive cutaneous squamous cell carcinoma. Clin. Cancer Res. 20, 6582–6592 (2014).
Li, Y.Y. et al. Genomic analysis of metastatic cutaneous squamous cell carcinoma. Clin. Cancer Res. 21, 1447–1456 (2015).
Stransky, N. et al. The mutational landscape of head and neck squamous cell carcinoma. Science 333, 1157–1160 (2011).
Agrawal, N. et al. Exome sequencing of head and neck squamous cell carcinoma reveals inactivating mutations in NOTCH1. Science 333, 1154–1157 (2011).
The Cancer Genome Atlas Research Network. Comprehensive genomic characterization of squamous cell lung cancers. Nature 489, 519–525 (2012).
Song, Y. et al. Identification of genomic alterations in oesophageal squamous cell cancer. Nature 509, 91–95 (2014).
Lin, D.C. et al. Genomic and molecular characterization of esophageal squamous cell carcinoma. Nat. Genet. 46, 467–473 (2014).
India Project Team of the International Cancer Genome Consortium. Mutational landscape of gingivo-buccal oral squamous cell carcinoma reveals new recurrently-mutated genes and molecular subgroups. Nat. Commun. 4, 2873 (2013).
Ojesina, A.I. et al. Landscape of genomic alterations in cervical carcinomas. Nature 506, 371–375 (2014).
Lin, D.C. et al. The genomic landscape of nasopharyngeal carcinoma. Nat. Genet. 46, 866–871 (2014).
Imielinski, M. et al. Mapping the hallmarks of lung adenocarcinoma with massively parallel sequencing. Cell 150, 1107–1120 (2012).
Wu, W. et al. HERC2 is an E3 ligase that targets BRCA1 for degradation. Cancer Res. 70, 6384–6392 (2010).
Toll, A. et al. MYC gene numerical aberrations in actinic keratosis and cutaneous squamous cell carcinoma. Br. J. Dermatol. 161, 1112–1118 (2009).
Al Bashir, S. et al. Cysteine-rich secretory protein 3 (CRISP3), ERG and PTEN define a molecular subtype of prostate cancer with implication to patients' prognosis. J. Hematol. Oncol. 7, 21 (2014).
Quintana, R.M. et al. A transposon-based analysis of gene mutations related to skin cancer development. J. Invest. Dermatol. 133, 239–248 (2013).
McFadden, D.G. et al. Genetic and clonal dissection of murine small cell lung carcinoma progression by genome sequencing. Cell 156, 1298–1311 (2014).
Dudgeon, C. et al. The evolution of thymic lymphomas in p53 knockout mice. Genes Dev. 28, 2613–2620 (2014).
Hennings, H. et al. FVB/N mice: an inbred strain sensitive to the chemical induction of squamous cell carcinomas in the skin. Carcinogenesis 14, 2353–2358 (1993).
Lapouge, G. et al. Skin squamous cell carcinoma propagating cells increase with tumour progression and invasiveness. EMBO J. 31, 4563–4575 (2012).
Miller, S.J. et al. Mouse skin is particularly susceptible to tumor initiation during early anagen of the hair cycle: possible involvement of hair follicle stem cells. J. Invest. Dermatol. 101, 591–594 (1993).
Blanpain, C., Lowry, W.E., Geoghegan, A., Polak, L. & Fuchs, E. Self-renewal, multipotency, and the existence of two cell populations within an epithelial stem cell niche. Cell 118, 635–648 (2004).
Li, H. & Durbin, R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25, 1754–1760 (2009).
Li, H. et al. The Sequence Alignment/Map format and SAMtools. Bioinformatics 25, 2078–2079 (2009).
McKenna, A. et al. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 20, 1297–1303 (2010).
Albers, C.A. et al. Dindel: accurate indel calls from short-read data. Genome Res. 21, 961–973 (2011).
Quinlan, A.R. & Hall, I.M. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics 26, 841–842 (2010).
Langmead, B. & Salzberg, S.L. Fast gapped-read alignment with Bowtie 2. Nat. Methods 9, 357–359 (2012).
Dees, N.D. et al. MuSiC: identifying mutational significance in cancer genomes. Genome Res. 22, 1589–1598 (2012).
Lawrence, M.S. et al. Mutational heterogeneity in cancer and the search for new cancer-associated genes. Nature 499, 214–218 (2013).
Farkash-Amar, S. et al. Global organization of replication time zones of the mouse genome. Genome Res. 18, 1562–1570 (2008).
Choi, Y., Sims, G.E., Murphy, S., Miller, J.R. & Chan, A.P. Predicting the functional effect of amino acid substitutions and indels. PLoS One 7, e46688 (2012).
Greenman, C. et al. Patterns of somatic mutation in human cancer genomes. Nature 446, 153–158 (2007).
Risso, D., Schwartz, K., Sherlock, G. & Dudoit, S. GC-content normalization for RNA-Seq data. BMC Bioinformatics 12, 480 (2011).
Cerami, E. et al. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2, 401–404 (2012).
Gao, J. et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci. Signal. 6, pl1 (2013).
Wang, K. et al. PennCNV: an integrated hidden Markov model designed for high-resolution copy number variation detection in whole-genome SNP genotyping data. Genome Res. 17, 1665–1674 (2007).
Beroukhim, R. et al. Assessing the significance of chromosomal aberrations in cancer: methodology and application to glioma. Proc. Natl. Acad. Sci. USA 104, 20007–20012 (2007).
Carter, S.L. et al. Absolute quantification of somatic DNA alterations in human cancer. Nat. Biotechnol. 30, 413–421 (2012).
Acknowledgements
C.B. is an investigator at Walloon Excellence in Life Sciences and Biotechnology (WELBIO). D.N. and M.L. are supported by Télévie and an EMBO long-term fellowship, respectively. The computational resources and services used in this work were provided by the Flemish Supercomputer Center (VSC), funded by the Hercules Foundation and the Flemish Government, Department of Economy, Science and Innovation (EWI). This work was supported by Fonds de la Recherche Scientifique (FNRS), Télévie, the Interuniversity Attraction Program (PAI), a research grant and an infrastructure grant from Fondation Contre le Cancer/Stichting tegen Kanker, Fondation ULB, Fond Yvonne Boël, Fond Gaston Ithier, the the Baillet Latour Foundation and the European Research Council (ERC).
Author information
Authors and Affiliations
Contributions
C.B., D.N. and D.L. designed the experiments. B.B. and D.L. performed bioinformatic analysis. D.N. and M.L. performed biological experiments. All authors performed data analysis. C.B., D.L. and D.N. wrote the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–4 (PDF 6667 kb)
Supplementary Data
Sequencing data and analysis (XLSX 8166 kb)
Rights and permissions
About this article
Cite this article
Nassar, D., Latil, M., Boeckx, B. et al. Genomic landscape of carcinogen-induced and genetically induced mouse skin squamous cell carcinoma. Nat Med 21, 946–954 (2015). https://doi.org/10.1038/nm.3878
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nm.3878
This article is cited by
-
Advances in cutaneous squamous cell carcinoma
Nature Reviews Cancer (2023)
-
Lymphatic Drainage System and Lymphatic Metastasis of Cancer Cells in the Mouse Esophagus
Digestive Diseases and Sciences (2023)
-
Notch signaling pathway: architecture, disease, and therapeutics
Signal Transduction and Targeted Therapy (2022)
-
Synthetic lethal kinases in Ras/p53 mutant squamous cell carcinoma
Oncogene (2022)
-
Cellular Heterogeneity and Plasticity of Skin Epithelial Cells in Wound Healing and Tumorigenesis
Stem Cell Reviews and Reports (2022)