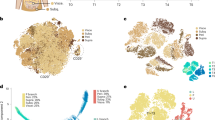
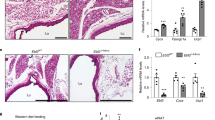
Abstract
Targeting brown adipose tissue (BAT) content or activity has therapeutic potential for treating obesity and the metabolic syndrome by increasing energy expenditure. However, both inter- and intra-individual differences contribute to heterogeneity in human BAT and potentially to differential thermogenic capacity in human populations. Here we generated clones of brown and white preadipocytes from human neck fat and characterized their adipogenic and thermogenic differentiation. We combined an uncoupling protein 1 (UCP1) reporter system and expression profiling to define novel sets of gene signatures in human preadipocytes that could predict the thermogenic potential of the cells once they were maturated. Knocking out the positive UCP1 regulators, PREX1 and EDNRB, in brown preadipocytes using CRISPR-Cas9 markedly abolished the high level of UCP1 in brown adipocytes differentiated from the preadipocytes. Finally, we were able to prospectively isolate adipose progenitors with great thermogenic potential using the cell surface marker CD29. These data provide new insights into the cellular heterogeneity in human fat and offer potential biomarkers for identifying thermogenically competent preadipocytes.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
Accession codes
References
Cannon, B. & Nedergaard, J. Brown adipose tissue: function and physiological significance. Physiol. Rev. 84, 277–359 (2004).
Schulz, T.J. & Tseng, Y.H. Brown adipose tissue: development, metabolism and beyond. Biochem. J. 453, 167–178 (2013).
Guerra, C., Koza, R.A., Yamashita, H., Walsh, K. & Kozak, L.P. Emergence of brown adipocytes in white fat in mice is under genetic control. Effects on body weight and adiposity. J. Clin. Invest. 102, 412–420 (1998).
Petrovic, N. et al. Chronic peroxisome proliferator-activated receptor gamma (PPARγ) activation of epididymally derived white adipocyte cultures reveals a population of thermogenically competent, UCP1-containing adipocytes molecularly distinct from classic brown adipocytes. J. Biol. Chem. 285, 7153–7164 (2010).
Harms, M. & Seale, P. Brown and beige fat: development, function and therapeutic potential. Nat. Med. 19, 1252–1263 (2013).
Nedergaard, J. & Cannon, B. The browning of white adipose tissue: some burning issues. Cell Metab. 20, 396–407 (2014).
Bartelt, A. et al. Brown adipose tissue activity controls triglyceride clearance. Nat. Med. 17, 200–205 (2011).
Stanford, K.I. et al. Brown adipose tissue regulates glucose homeostasis and insulin sensitivity. J. Clin. Invest. 123, 215–223 (2013).
Nedergaard, J., Bengtsson, T. & Cannon, B. Unexpected evidence for active brown adipose tissue in adult humans. Am. J. Physiol. Endocrinol. Metab. 293, E444–E452 (2007).
Cypess, A.M. et al. Identification and importance of brown adipose tissue in adult humans. N. Engl. J. Med. 360, 1509–1517 (2009).
van Marken Lichtenbelt, W.D. et al. Cold-activated brown adipose tissue in healthy men. N. Engl. J. Med. 360, 1500–1508 (2009).
Virtanen, K.A. et al. Functional brown adipose tissue in healthy adults. N. Engl. J. Med. 360, 1518–1525 (2009).
Saito, M. et al. High incidence of metabolically active brown adipose tissue in healthy adult humans: effects of cold exposure and adiposity. Diabetes 58, 1526–1531 (2009).
Zingaretti, M.C. et al. The presence of UCP1 demonstrates that metabolically active adipose tissue in the neck of adult humans truly represents brown adipose tissue. FASEB J. 23, 3113–3120 (2009).
Himms-Hagen, J. et al. Effect of CL-316,243, a thermogenic β3-agonist, on energy balance and brown and white adipose tissues in rats. Am. J. Physiol. 266, R1371–R1382 (1994).
Yoneshiro, T. et al. Recruited brown adipose tissue as an antiobesity agent in humans. J. Clin. Invest. 123, 3404–3408 (2013).
van der Lans, A.A. et al. Cold acclimation recruits human brown fat and increases nonshivering thermogenesis. J. Clin. Invest. 123, 3395–3403 (2013).
Ouellet, V. et al. Brown adipose tissue oxidative metabolism contributes to energy expenditure during acute cold exposure in humans. J. Clin. Invest. 122, 545–552 (2012).
Wu, J. et al. Beige adipocytes are a distinct type of thermogenic fat cell in mouse and human. Cell 150, 366–376 (2012).
Cypess, A.M. et al. Anatomical localization, gene expression profiling and functional characterization of adult human neck brown fat. Nat. Med. 19, 635–639 (2013).
Lidell, M.E. et al. Evidence for two types of brown adipose tissue in humans. Nat. Med. 19, 631–634 (2013).
Jespersen, N.Z. et al. A classical brown adipose tissue mRNA signature partly overlaps with brite in the supraclavicular region of adult humans. Cell Metab. 17, 798–805 (2013).
Schulz, T.J. et al. Identification of inducible brown adipocyte progenitors residing in skeletal muscle and white fat. Proc. Natl. Acad. Sci. USA 108, 143–148 (2011).
Lee, Y.H., Petkova, A.P., Mottillo, E.P. & Granneman, J.G. In vivo identification of bipotential adipocyte progenitors recruited by β3-adrenoceptor activation and high-fat feeding. Cell Metab. 15, 480–491 (2012).
Berry, R. & Rodeheffer, M.S. Characterization of the adipocyte cellular lineage in vivo. Nat. Cell Biol. 15, 302–308 (2013).
Wang, W. et al. Ebf2 is a selective marker of brown and beige adipogenic precursor cells. Proc. Natl. Acad. Sci. USA 111, 14466–14471 (2014).
Tchkonia, T. et al. Fat depot–specific characteristics are retained in strains derived from single human preadipocytes. Diabetes 55, 2571–2578 (2006).
Whittle, A.J. et al. BMP8B increases brown adipose tissue thermogenesis through both central and peripheral actions. Cell 149, 871–885 (2012).
Tseng, Y.H. et al. New role of bone morphogenetic protein 7 in brown adipogenesis and energy expenditure. Nature 454, 1000–1004 (2008).
Seale, P. et al. Transcriptional control of brown fat determination by PRDM16. Cell Metab. 6, 38–54 (2007).
Timmons, J.A. et al. Myogenic gene expression signature establishes that brown and white adipocytes originate from distinct cell lineages. Proc. Natl. Acad. Sci. USA 104, 4401–4406 (2007).
Welch, H.C. et al. P-Rex1, a PtdIns(3,4,5)P3- and Gβγ-regulated guanine-nucleotide exchange factor for Rac. Cell 108, 809–821 (2002).
Cheung, J. et al. Identification of the human cortactin-binding protein-2 gene from the autism candidate region at 7q31. Genomics 78, 7–11 (2001).
Zhang, S.X. et al. Identification of direct serum-response factor gene targets during Me2SO-induced P19 cardiac cell differentiation. J. Biol. Chem. 280, 19115–19126 (2005).
Yamada, Y. et al. Cloning and functional characterization of a family of human and mouse somatostatin receptors expressed in brain, gastrointestinal tract and kidney. Proc. Natl. Acad. Sci. USA 89, 251–255 (1992).
Kikkawa, T. et al. Dmrta1 regulates proneural gene expression downstream of Pax6 in the mammalian telencephalon. Genes Cells 18, 636–649 (2013).
Garciafigueroa, D.Y., Klei, L.R., Ambrosio, F. & Barchowsky, A. Arsenic-stimulated lipolysis and adipose remodeling is mediated by G-protein–coupled receptors. Toxicol. Sci. 134, 335–344 (2013).
Chen, T.Y. et al. Endogenous n-3 polyunsaturated fatty acids (PUFAs) mitigate ovariectomy-induced bone loss by attenuating bone marrow adipogenesis in FAT1 transgenic mice. Drug Des. Devel. Ther. 7, 545–552 (2013).
Behjati, S. et al. Recurrent PTPRB and PLCG1 mutations in angiosarcoma. Nat. Genet. 46, 376–379 (2014).
Takada, Y., Ye, X. & Simon, S. The integrins. Genome Biol. 8, 215 (2007).
Margadant, C., Monsuur, H.N., Norman, J.C. & Sonnenberg, A. Mechanisms of integrin activation and trafficking. Curr. Opin. Cell Biol. 23, 607–614 (2011).
Balamatsias, D. et al. Identification of P-Rex1 as a novel Rac1-guanine nucleotide exchange factor (GEF) that promotes actin remodeling and GLUT4 protein trafficking in adipocytes. J. Biol. Chem. 286, 43229–43240 (2011).
Lewis, J.P. et al. Analysis of candidate genes on chromosome 20q12–13.1 reveals evidence for BMI-mediated association of PREX1 with type 2 diabetes in European Americans. Genomics 96, 211–219 (2010).
Wu-Wong, J.R., Berg, C.E. & Dayton, B.D. Endothelin-stimulated glucose uptake: effects of intracellular Ca2+, cAMP and glucosamine. Clin. Sci. 103 (suppl. 48), 418S–423S (2002).
Juan, C.C. et al. Effect of endothelin-1 on lipolysis in rat adipocytes. Obesity (Silver Spring) 14, 398–404 (2006).
Shinoda, K. et al. Genetic and functional characterization of clonally derived adult human brown adipocytes. Nat. Med. 21, 389–394 (2015).
Gierloff, M. et al. Adipogenic differentiation potential of rat adipose tissue–derived subpopulations of stromal cells. J. Plast. Reconstr. Aesthet. Surg. 67, 1427–1435 (2014).
Farnier, C. et al. The signaling pathway for β1-integrin/ERKs is involved in the adaptation of adipocyte functions to cell size. Ann. NY Acad. Sci. 973, 594–597 (2002).
Kawaguchi, N. et al. ADAM12 induces actin cytoskeleton and extracellular matrix reorganization during early adipocyte differentiation by regulating β1 integrin function. J. Cell Sci. 116, 3893–3904 (2003).
Irizarry, R.A. et al. Exploration, normalization, and summaries of high-density oligonucleotide array probe level data. Biostatistics 4, 249–264 (2003).
Benjamin, Y. et al. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. R. Statist. Soc. B 57, 289–300 (1995).
Acknowledgements
This work was supported in part by US National Institutes of Health (NIH) grants R01DK077097 (to Y.-H.T.), R01DK099511 (to L.J.G.), K23DK081604 (to A.M.C.) and P30DK036836 (to Joslin Diabetes Center's Diabetes Research Center, DRC) from the National Institute of Diabetes and Digestive and Kidney Diseases, a sponsored research grant from Chugai Pharmaceutical Co. (to Y.-H.T. and A.M.C.), a research grant from the American Diabetes Association (ADA 7-12-BS-191, to Y.-H.T.), the Intramural Research Program of the National Institute of Diabetes and Digestive and Kidney Diseases (NIDDK), and by funding from the Harvard Stem Cell Institute (to Y.-H.T.). M.D.L. was supported by NIH fellowships (T32DK007260 and F32DK102320). We thank M.-E. Patti and K. Hughes of the Advanced Genomics and Genetics Core of Joslin's DRC for advice and expert technical assistance. The authors thank Stryker Regenerative Medicine (Hopkinton, Massachusetts) for the generous gift of recombinant BMP7.
Author information
Authors and Affiliations
Contributions
The study was designed by Y.-H.T., R.X., M.D.L. and A.M.C. The manuscript was written by Y.-H.T., M.D.L., R.X. and J.M.D. R.X. performed the majority of the experiments. M.D.L. did the time-lapse imaging, IVIS scanning and FACS. J.M.D. analyzed microarray data. F.S. performed bioenergetics analyses in knockout cells. T.J.S. and H.Z. established the method of isolation, immortalization and differentiation of human fat progenitors. T.L.H. did the human cell implantation and gene expression microarrays. K.L.T. provided assistance with the Seahorse bioanalyzer. Y.L. provided research assistance. H.T. and L.J.G. helped with fuel utilization experiments. A.M.C., L.S.W. and A.P.W. collected human fat samples. M.S.L. and L.L.R. helped with the time-lapse imaging. All authors contributed to editing the manuscript.
Corresponding author
Ethics declarations
Competing interests
A.M.C. and Y.-H.T. are recipients of a sponsored research grant and licensing payments from Chugai Pharmaceutical Co., Ltd through Joslin Diabetes Center.
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–9 & Supplementary Tables 1–5 (PDF 1480 kb)
Rights and permissions
About this article
Cite this article
Xue, R., Lynes, M., Dreyfuss, J. et al. Clonal analyses and gene profiling identify genetic biomarkers of the thermogenic potential of human brown and white preadipocytes. Nat Med 21, 760–768 (2015). https://doi.org/10.1038/nm.3881
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nm.3881
This article is cited by
-
TMEM135 links peroxisomes to the regulation of brown fat mitochondrial fission and energy homeostasis
Nature Communications (2023)
-
The serotonin transporter sustains human brown adipose tissue thermogenesis
Nature Metabolism (2023)
-
A non-coding variant linked to metabolic obesity with normal weight affects actin remodelling in subcutaneous adipocytes
Nature Metabolism (2023)
-
Browning of the white adipose tissue regulation: new insights into nutritional and metabolic relevance in health and diseases
Nutrition & Metabolism (2022)
-
Adipocyte Gq signaling is a regulator of glucose and lipid homeostasis in mice
Nature Communications (2022)