Abstract
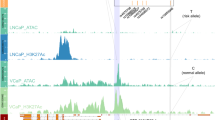
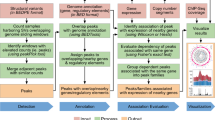
The vast majority of disease-associated single-nucleotide polymorphisms (SNPs) mapped by genome-wide association studies (GWASs) are located in the non-protein-coding genome, but establishing the functional and mechanistic roles of these sequence variants has proven challenging. Here we describe a general pipeline in which candidate functional SNPs are first evaluated by fine mapping, epigenomic profiling, and epigenome editing, and then interrogated for causal function by using genome editing to create isogenic cell lines followed by phenotypic characterization. To validate this approach, we analyzed the 6q22.1 prostate cancer risk locus and identified rs339331 as the top-scoring SNP. Epigenome editing confirmed that the rs339331 region possessed regulatory potential. By using transcription activator-like effector nuclease (TALEN)-mediated genome editing, we created a panel of isogenic 22Rv1 prostate cancer cell lines representing all three genotypes (TT, TC, CC) at rs339331. Introduction of the 'T' risk allele increased transcription of the regulatory factor 6 (RFX6) gene, increased homeobox B13 (HOXB13) binding at the rs339331 region, and increased deposition of the enhancer-associated H3K4me2 histone mark at the rs339331 region compared to lines homozygous for the 'C' protective allele. The cell lines also differed in cellular morphology and adhesion, and pathway analysis of differentially expressed genes suggested an influence of androgens. In summary, we have developed and validated a widely accessible approach that can be used to establish functional causality for noncoding sequence variants identified by GWASs.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
Accession codes
References
Hindorff, L.A. et al. Potential etiologic and functional implications of genome-wide association loci for human diseases and traits. Proc. Natl. Acad. Sci. USA 106, 9362–9367 (2009).
Cookson, W., Liang, L., Abecasis, G., Moffatt, M. & Lathrop, M. Mapping complex disease traits with global gene expression. Nat. Rev. Genet. 10, 184–194 (2009).
Nicolae, D.L. et al. Trait-associated SNPs are more likely to be eQTLs: annotation to enhance discovery from GWAS. PLoS Genet. 6, e1000888 (2010).
The ENCODE Project Consortium. An integrated encyclopedia of DNA elements in the human genome. Nature 489, 57–74 (2012).
Maurano, M.T. et al. Systematic localization of common disease-associated variation in regulatory DNA. Science 337, 1190–1195 (2012).
Bauer, D.E. et al. An erythroid enhancer of BCL11A subject to genetic variation determines fetal hemoglobin level. Science 342, 253–257 (2013).
Ochiai, H. et al. TALEN-mediated single-base-pair editing identification of an intergenic mutation upstream of BUB1B as causative of PCS (MVA) syndrome. Proc. Natl. Acad. Sci. USA 111, 1461–1466 (2014).
Albert, F.W. & Kruglyak, L. The role of regulatory variation in complex traits and disease. Nat. Rev. Genet. 16, 197–212 (2015).
Knight, J.C. Approaches for establishing the function of regulatory genetic variants involved in disease. Genome Med 6, 92 (2014).
Hsu, P.D., Lander, E.S. & Zhang, F. Development and applications of CRISPR-Cas9 for genome engineering. Cell 157, 1262–1278 (2014).
Takata, R. et al. Genome-wide association study identifies five new susceptibility loci for prostate cancer in the Japanese population. Nat. Genet. 42, 751–754 (2010).
Huang, Q. et al. A prostate cancer susceptibility allele at 6q22 increases RFX6 expression by modulating HOXB13 chromatin binding. Nat. Genet. 46, 126–135 (2014).
Han, Y. et al. Integration of multiethnic fine-mapping and genomic annotation to prioritize candidate functional SNPs at prostate cancer susceptibility regions. Hum. Mol. Genet. 24, 5603–5618 (2015).
Joung, J.K. & Sander, J.D. TALENs: a widely applicable technology for targeted genome editing. Nat. Rev. Mol. Cell Biol. 14, 49–55 (2013).
Mendenhall, E.M. et al. Locus-specific editing of histone modifications at endogenous enhancers. Nat. Biotechnol. 31, 1133–1136 (2013).
Maeder, M.L. et al. Robust, synergistic regulation of human gene expression using TALE activators. Nat. Methods 10, 243–245 (2013).
Reyon, D. et al. FLASH assembly of TALENs for high-throughput genome editing. Nat. Biotechnol. 30, 460–465 (2012).
Kim, Y., Kweon, J. & Kim, J.S. TALENs and ZFNs are associated with different mutation signatures. Nat. Methods 10, 185 (2013).
Joung, J.K. Unwanted mutations: Standards needed for gene-editing errors. Nature 523, 158 (2015).
Guilinger, J.P. et al. Broad specificity profiling of TALENs results in engineered nucleases with improved DNA-cleavage specificity. Nat. Methods 11, 429–435 (2014).
Hockemeyer, D. et al. Genetic engineering of human pluripotent cells using TALE nucleases. Nat. Biotechnol. 29, 731–734 (2011).
Kim, D., et al. Digenome-seq: genome-wide profiling of CRISPR-Cas9 off-target effects in human cells. Nat. Methods 12, 237–243 (2015).
Wang, X. et al. Unbiased detection of off-target cleavage by CRISPR-Cas9 and TALENs using integrase-defective lentiviral vectors. Nat. Biotechnol. 33, 175–178 (2015).
Tsai, S.Q. et al. GUIDE-seq enables genome-wide profiling of off-target cleavage by CRISPR-Cas nucleases. Nat. Biotechnol. 33, 187–197 (2015).
Frock, R.L. et al. Genome-wide detection of DNA double-stranded breaks induced by engineered nucleases. Nat. Biotechnol. 33, 179–186 (2015).
Duggan, D. et al. Two genome-wide association studies of aggressive prostate cancer implicate putative prostate tumor suppressor gene DAB2IP. J. Natl. Cancer Inst. 99, 1836–1844 (2007).
Schumacher, F.R. et al. Genome-wide association study identifies new prostate cancer susceptibility loci. Hum. Mol. Genet. 20, 3867–3875 (2011).
Gohagan, J.K., Prorok, P.C., Hayes, R.B. & Kramer, B.S. The Prostate, Lung, Colorectal and Ovarian (PLCO) Cancer Screening Trial of the National Cancer Institute: history, organization, and status. Control. Clin. Trials 21, 251S–272S (2000).
Kolonel, L.N. et al. A multiethnic cohort in Hawaii and Los Angeles: baseline characteristics. Am. J. Epidemiol. 151, 346–357 (2000).
Cook, M.B. et al. A genome-wide association study of prostate cancer in West African men. Hum. Genet. 133, 509–521 (2014).
Cheng, I. et al. Evaluating genetic risk for prostate cancer among Japanese and Latinos. Cancer Epidemiol. Biomarkers Prev. 21, 2048–2058 (2012).
Akamatsu, S. et al. Common variants at 11q12, 10q26 and 3p11.2 are associated with prostate cancer susceptibility in Japanese. Nat. Genet. 44, 426–429 (2012).
Howie, B.N., Donnelly, P. & Marchini, J. A flexible and accurate genotype imputation method for the next generation of genome-wide association studies. PLoS Genet. 5, e1000529 (2009).
Pruim, R.J. et al. LocusZoom: regional visualization of genome-wide association scan results. Bioinformatics 26, 2336–2337 (2010).
Reyon, D. et al. Engineering customized TALE nucleases (TALENs) and TALE transcription factors by fast ligation-based automatable solid-phase high-throughput (FLASH) assembly. Curr. Protoc. Mol. Biol. 103, 12.16.1–12.16.18 (2013).
Chen, F. et al. High-frequency genome editing using ssDNA oligonucleotides with zinc-finger nucleases. Nat. Methods 8, 753–755 (2011).
R Core Team. R: A language and environment for statistical computing. (R Foundation for Statistical Computing, 2015).
Kim, D. et al. TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 14, R36 (2013).
Acknowledgements
M.L.F. and J.K.J. were supported by US National Institutes of Health (NIH) grant no. R01 GM107427; J.K.J. was supported by a NIH Director's Pioneer Award (DP1 GM105378) and The Jim and Ann Orr Massachusetts General Hospital Research Scholar Award; M.L.F. is supported by the Prostate Cancer Foundation (Challenge Award), the US NIH grant no. R01CA193910, and the H.L. Snyder Medical Foundation. The scientific development and funding for this project were in part supported by the US National Cancer Institute GAME-ON Post-GWAS Initiative (U19CA148112 and U19CA148537). K.L. is supported by a K99/R00 grant from the National Cancer Institute (grant no. 1K99CA184415-01). I.C. is supported by grants from the Hungarian National Research, Development and Innovation Office (KMR-12-1-2012-0216) and the Hungarian National Research Fund (OTKA103244). Z.S. is supported by the Breast Cancer Research Foundation and the Széchenyi Progam, Hungary (KTIA_NAP_13-2014-0021). This project was also supported by a Program Project Development Grant from the Ovarian Cancer Research Fund (K.L. and S.A.G). We thank the Dana-Farber Cancer Institute Molecular Biology Core Facility for Sanger sequencing and Illumina high-throughput sequencing. We thank C. Nicolet at the University of Southern California Epigenome Center Core for RNA-seq services and M. Li at the University of Southern California Norris Medical Library Bioinformatics Center, who provided assistance with the analysis of RNA-seq data.
Author information
Authors and Affiliations
Consortia
Contributions
S.S., K.L. and Y.F. designed and performed experiments, J.K.J. and M.L.F. designed experiments, R.T.C., J.-H.S., R.L., V.T., M.C. and M.P. performed experiments, S.S., I.C. and M.L.F. developed the sequencing pipeline, S.S., K.L., Y.F., Y.H., Q.L., I.C., Z.T.H. and N.S. analyzed the data, S.S., K.L., I.C., J.K.J. and M.L.F. wrote the manuscript, S.S., Y.F., R.T.C., S.A.G., J.K.J. and M.L.F. revised the manuscript, C.H., Z.S., Z.T.H. and S.A.G. provided technical support and conceptual advice. The GAME-ON/ELLIPSE Consortium provided early access to fine-mapping data.
Corresponding authors
Ethics declarations
Competing interests
J.K.J. is a consultant for Horizon Discovery. J.K.J. has financial interests in Editas Medicine, Hera Testing Laboratories, Poseida Therapeutics, and Transposagen Biopharmaceuticals. J.K.J.'s interests were reviewed and are managed by Massachusetts General Hospital and Partners HealthCare in accordance with their conflict of interest policies.
Additional information
A complete list of all consortium members is provided in the Supplementary Note.
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–10 and Supplementary Note (PDF 9987 kb)
Supplementary Table 1
The most associated markers for the 6q22.1 prostate risk locus (XLSX 20 kb)
Supplementary Table 2
Description and study design of the studies included in the meta-analysis. (XLSX 18 kb)
Supplementary Table 3
Identified allele variants (459) and their fraquencies. Deleted base represented by “x” (XLSX 16 kb)
Supplementary Table 4
Differentially expressed transcripts between rs339331 isogenic series (XLSX 13 kb)
Supplementary Table 5
Oligonucleotide seqences (XLSX 12 kb)
Rights and permissions
About this article
Cite this article
Spisák, S., Lawrenson, K., Fu, Y. et al. CAUSEL: an epigenome- and genome-editing pipeline for establishing function of noncoding GWAS variants. Nat Med 21, 1357–1363 (2015). https://doi.org/10.1038/nm.3975
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nm.3975
This article is cited by
-
GATA2 co-opts TGFβ1/SMAD4 oncogenic signaling and inherited variants at 6q22 to modulate prostate cancer progression
Journal of Experimental & Clinical Cancer Research (2023)
-
Construction of lncRNA/Pseudogene-miRNA Network Based on In Silico Approaches for Glycolysis Pathway to Identify Prostate Adenocarcinoma-Related Potential Biomarkers
Applied Biochemistry and Biotechnology (2023)
-
A biallelic multiple nucleotide length polymorphism explains functional causality at 5p15.33 prostate cancer risk locus
Nature Communications (2023)
-
HOXB13 suppresses de novo lipogenesis through HDAC3-mediated epigenetic reprogramming in prostate cancer
Nature Genetics (2022)
-
Augmenting and directing long-range CRISPR-mediated activation in human cells
Nature Methods (2021)