Abstract
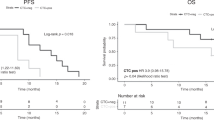
Assessment of KRAS status is mandatory in patients with metastatic colorectal cancer (mCRC) before applying targeted therapy. We describe here a blinded prospective study to compare KRAS and BRAF mutation status data obtained from the analysis of tumor tissue by routine gold-standard methods and of plasma DNA using a quantitative PCR–based method specifically designed to analyze circulating cell-free DNA (cfDNA). The mutation status was determined by both methods from 106 patient samples. cfDNA analysis showed 100% specificity and sensitivity for the BRAF V600E mutation. For the seven tested KRAS point mutations, the method exhibited 98% specificity and 92% sensitivity with a concordance value of 96%. Mutation load, expressed as the proportion of mutant alleles in cfDNA, was highly variable (0.5–64.1%, median 10.5%) among mutated samples. CfDNA was detected in 100% of patients with mCRC. This study shows that liquid biopsy through cfDNA analysis could advantageously replace tumor-section analysis and expand the scope of personalized medicine for patients with cancer.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout


Similar content being viewed by others
References
Lièvre, A. et al. KRAS mutation status is predictive of response to cetuximab therapy in colorectal cancer. Cancer Res. 66, 3992–3995 (2006).
Amado, R.G. et al. Wild-type KRAS is required for panitumumab efficacy in patients with metastatic colorectal cancer. J. Clin. Oncol. 26, 1626–1634 (2008).
Karapetis, C.S. et al. K-ras mutations and benefit from cetuximab in advanced colorectal cancer. N. Engl. J. Med. 359, 1757–1765 (2008).
Bando, H. et al. KRAS mutations detected by the amplification refractory mutation system-Scorpion assays strongly correlate with therapeutic effect of cetuximab. Br. J. Cancer 105, 403–406 (2011).
Pritchard, C.C. & Grady, W.M. Colorectal cancer molecular biology moves into clinical practice. Gut 60, 116–129 (2011).
Van Cutsem, E. et al. Cetuximab plus irinotecan, fluorouracil, and leucovorin as first-line treatment for metastatic colorectal cancer: updated analysis of overall survival according to tumor KRAS and BRAF mutation status. J. Clin. Oncol. 29, 2011–2019 (2011).
Laurent-Puig, P. et al. Analysis of PTEN, BRAF, and EGFR status in determining benefit from cetuximab therapy in wild-type KRAS metastatic colon cancer. J. Clin. Oncol. 27, 5924–5930 (2009).
Bibeau, F. et al. Impact of FcγRIIa-FcγRIIIa polymorphisms and KRAS mutations on the clinical outcome of patients with metastatic colorectal cancer treated with cetuximab plus irinotecan. J. Clin. Oncol. 27, 1122–1129 (2009).
Kobunai, T., Watanabe, T., Yamamoto, Y. & Eshima, K. The frequency of KRAS mutation detection in human colon carcinoma is influenced by the sensitivity of assay methodology: a comparison between direct sequencing and real-time PCR. Biochem. Biophys. Res. Commun. 395, 158–162 (2010).
Pinto, P. et al. Comparison of methodologies for KRAS mutation detection in metastatic colorectal cancer. Cancer Genetics 204, 439–446 (2011).
Weichert, W. et al. KRAS genotyping of paraffin-embedded colorectal cancer tissue in routine diagnostics: comparison of methods and impact of histology. J. Mol. Diagn. 12, 35–42 (2010).
Boissière-Michot, F. et al. KRAS genotyping in rectal adenocarcinoma specimens with low tumor cellularity after neoadjuvant treatment. Mod. Pathol. 25, 731–739 (2012).
Artru, P. et al. Review of the current status of KRAS mutation testing in France in 2011: The Flash-KRAS study. J. Clin. Oncol. 30 (suppl.), e14129 (2012).
Stroun, M., Anker, P., Lyautey, J., Lederrey, C. & Maurice, P.A. isolation and characterization of dna from the plasma of cancer-patients. Eur. J. Cancer Clin. Oncol. 23, 707–712 (1987).
Lecomte, T. et al. Detection of free-circulating tumor-associated DNA in plasma of colorectal cancer patients and its association with prognosis. Int. J. Cancer 100, 542–548 (2002).
Lefebure, B. et al. Prognostic value of circulating mutant DNA in unresectable metastatic colorectal cancer. Ann. Surg. 251, 275–280 (2010).
Thierry, A.R. et al. Origin and quantification of circulating DNA in mice with human colorectal cancer xenografts. Nucleic Acids Res. 38, 6159–6175 (2010).
Schwarzenbach, H., Hoon, D.S.B. & Pantel, K. Cell-free nucleic acids as biomarkers in cancer patients. Nat. Rev. Cancer 11, 426–437 (2011).
Mead, R., Duku, M., Bhandari, P. & Cree, I.A. Circulating tumour markers can define patients with normal colons, benign polyps, and cancers. Br. J. Cancer 105, 239–245 (2011).
Diehl, F. et al. Circulating mutant DNA to assess tumor dynamics. Nat. Med. 14, 985–990 (2008).
Mouliere, F. et al. High fragmentation characterizes tumour-derived circulating DNA. PLoS One 6, e23418 (2011).
Mouliere, F. & Thierry, A.R. The importance of examining the proportion of circulating DNA originating from tumor, microenvironment and normal cells in colorectal cancer patients. Expert Opin. Biol. Ther. 12 (suppl. 1), S209–S215 (2012).
Trevisiol, C. et al. Prognostic value of circulating KRAS2 gene mutations in colorectal cancer with distant metastases. Int. J. Biol. Markers 21, 223–228 (2006).
Ryan, B.M. et al. A prospective study of circulating mutant KRAS2 in the serum of patients with colorectal neoplasia: strong prognostic indicator in postoperative follow up. Gut 52, 101–108 (2003).
Yen, L.C. et al. Detection of KRAS oncogene in peripheral blood as a predictor of the response to cetuximab plus chemotherapy in patients with metastatic colorectal cancer. Clin. Cancer Res. 15, 4508–4513 (2009).
Spindler, K.L.G., Pallisgaard, N., Vogelius, I. & Jakobsen, A. quantitative cell-free DNA, KRAS, and BRAF mutations in plasma from patients with metastatic colorectal cancer during treatment with cetuximab and irinotecan. Clin. Cancer Res. 18, 1177–1185 (2012).
Kim, M.J. et al. Different metastatic pattern according to the KRAS mutational status and site-specific discordance of KRAS status in patients with colorectal cancer. BMC Cancer 12, 347 (2012).
Knijn, N. et al. KRAS mutation analysis: a comparison between primary tumours and matched liver metastases in 305 colorectal cancer patients. Br. J. Cancer 104, 1020–1026 (2011).
Oltedal, S. et al. Heterogeneous distribution of K-ras mutations in primary colon carcinomas: implications for EGFR-directed therapy. Int. J. Colorectal Dis. 26, 1271–1277 (2011).
Gonzalez de Castro, D. et al. A comparison of three methods for detecting KRAS mutations in formalin-fixed colorectal cancer specimens. Br. J. Cancer 107, 345–351 (2012).
Solassol, J. et al. KRAS mutation detection in paired frozen and formalin-fixed paraffin-embedded (FFPE) colorectal cancer tissues. Int. J. Mol. Sci. 12, 3191–3204 (2011).
Lièvre, A. et al. KRAS mutations as an independent prognostic factor in patients with advanced colorectal cancer treated with cetuximab. J. Clin. Oncol. 26, 374–379 (2008).
Misale, S. et al. Emergence of KRAS mutations and acquired resistance to anti-EGFR therapy in colorectal cancer. Nature 486, 532–536 (2012).
Diaz, L.A. et al. The molecular evolution of acquired resistance to targeted EGFR blockade in colorectal cancers. Nature 486, 537–540 (2012).
Diehl, F. et al. Analysis of mutations in DNA isolated from plasma and stool of colorectal cancer patients. Gastroenterology 135, 489–498 (2008).
Leary, R.J. et al. Development of personalized tumor biomarkers using massively parallel sequencing. Sci. Transl. Med. 2, 20ra14 (2010).
Board, R.E. et al. Detection of PIK3CA mutations in circulating free DNA in patients with breast cancer. Breast Cancer Res. Treat. 120, 461–467 (2010).
West, N.P. et al. The proportion of tumour cells is an independent predictor for survival in colorectal cancer patients. Br. J. Cancer 102, 1519–1523 (2010).
Soh, J. et al. Oncogene mutations, copy number gains and mutant allele specific imbalance (MASI) frequently occur together in tumor cells. PLoS ONE 4, e7464 (2009).
El Messaoudi, S., Rolet, F., Mouliere, F. & Thierry, A.R. Circulating cell free DNA: Preanalytical considerations. Clin. Chim. Acta 424, 222–230 (2013).
Bustin, S.A. et al. The MIQE guidelines: minimum information for publication of quantitative real-time PCR experiments. Clin. Chem. 55, 611–622 (2009).
McShane, L.M. et al. REporting recommendations for tumor MARKer prognostic studies (REMARK). Nat. Clin. Pract. Oncol. 2, 416–422 (2005).
Acknowledgements
We thank J. Robert, J. Dubois, A. Lievre, C. Theillet, S. Du Manoir, C. Sardet, D. Tousch and R. Gimbaud for helpful discussions. We would like to thank L. Kamraoui, A. Laybats, J.-Y. Cance and M. Cavalier for excellent technical assistance. F. Mouliere is supported by a grant from CNRS and the Region of Languedoc-Roussillon (CNRS044406). The study was funded by grants from the Fondation ARC pour la Recherche sur le Cancer and the Association GEFLUC (DCMLP10-184, France). We thank Bio-Rad for the qPCR thermocycler free loan. A.R.T. is supported by INSERM.
Author information
Authors and Affiliations
Contributions
A.R.T. obtained funding, designed the study, interpreted experiments and wrote the manuscript. M.M., D.P., F.B., E.L.-C., M.D.R., P.-J.L., F. Mouliere and M.Y. designed the study. F. Mouliere, S.E.M. and F.R. performed experiments and analyzed the data. F. Mouliere, S.E.M. and A.R.T. prepared the manuscript. C.G., B.R., S.E.M., E.L.-C., M.D.R., M.M., D.P. and M.Y. critically revised the manuscript. C.G., B.R. and F. Molina gave conceptual advice and assisted in the design of experiments. C.M. designed the study, acquired all of the clinical data and performed the statistical analysis. E.L.-C., B.G., P.D., M.N., V.L. and A.-S.J. collected blood samples and acquired the clinical data. F. Mouliere, S.E.M. and A.R.T. developed the Intplex assay. C.G., B.R., F. Mouliere, F. Molina, S.E.M., E.L.-C., M.D.R., M.M., D.P., F.B., P.-J.L., C.M. and M.Y. interpreted the experiments. All authors approved the content of the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–4, Supplementary Tables 1–4 and Supplementary Methods (PDF 1516 kb)
Rights and permissions
About this article
Cite this article
Thierry, A., Mouliere, F., El Messaoudi, S. et al. Clinical validation of the detection of KRAS and BRAF mutations from circulating tumor DNA. Nat Med 20, 430–435 (2014). https://doi.org/10.1038/nm.3511
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nm.3511
This article is cited by
-
Clinical application and detection techniques of liquid biopsy in gastric cancer
Molecular Cancer (2023)
-
High-throughput extraction on a dynamic solid phase for low-abundance biomarker isolation from biological samples
Microsystems & Nanoengineering (2023)
-
A straightforward method to quantify circulating mRNAs as biomarkers of colorectal cancer
Scientific Reports (2023)
-
Screening for a practical method to monitor the status of patients with metastatic bladder cancer at the circulating cell-gene level
Scientific Reports (2023)
-
Combining the amplification refractory mutation system and high-resolution melting analysis for KRAS mutation detection in clinical samples
Analytical and Bioanalytical Chemistry (2023)