Abstract
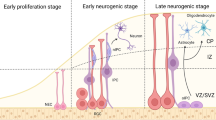
The exploration of brain epigenomes, which consist of various types of DNA methylation and covalent histone modifications, is providing new and unprecedented insights into the mechanisms of neural development, neurological disease and aging. Traditionally, chromatin defects in the brain were considered static lesions of early development that occurred in the context of rare genetic syndromes, but it is now clear that mutations and maladaptations of the epigenetic machinery cover a much wider continuum that includes adult-onset neurodegenerative disease. Here, we describe how recent advances in neuroepigenetics have contributed to an improved mechanistic understanding of developmental and degenerative brain disorders, and we discuss how they could influence the development of future therapies for these conditions.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout

Katie Vicari

Katie Vicari
Similar content being viewed by others
References
Numata, S. et al. DNA methylation signatures in development and aging of the human prefrontal cortex. Am. J. Hum. Genet. 90, 260–272 (2012).
Siegmund, K.D. et al. DNA methylation in the human cerebral cortex is dynamically regulated throughout the life span and involves differentiated neurons. PLoS One 2, e895 (2007).
Hernandez, D.G. et al. Distinct DNA methylation changes highly correlated with chronological age in the human brain. Hum. Mol. Genet. 20, 1164–1172 (2011).
Cheung, I. et al. Developmental regulation and individual differences of neuronal H3K4me3 epigenomes in the prefrontal cortex. Proc. Natl. Acad. Sci. USA 107, 8824–8829 (2010).
Klein, C.J. et al. Mutations in DNMT1 cause hereditary sensory neuropathy with dementia and hearing loss. Nat. Genet. 43, 595–600 (2011).
Winkelmann, J. et al. Mutations in DNMT1 cause autosomal dominant cerebellar ataxia, deafness and narcolepsy. Hum. Mol. Genet. 21, 2205–2210 (2012).
Peter, C.J. & Akbarian, S. Balancing histone methylation activities in psychiatric disorders. Trends Mol. Med. 17, 372–379 (2011).
Chuang, D.M., Leng, Y., Marinova, Z., Kim, H.J. & Chiu, C.T. Multiple roles of HDAC inhibition in neurodegenerative conditions. Trends Neurosci. 32, 591–601 (2009).
Baltan, S., Murphy, S.P., Danilov, C.A., Bachleda, A. & Morrison, R.S. Histone deacetylase inhibitors preserve white-matter structure and function during ischemia by conserving ATP and reducing excitotoxicity. J. Neurosci. 31, 3990–3999 (2011).
Fischer, A., Sananbenesi, F., Mungenast, A. & Tsai, L.H. Targeting the correct HDAC(s) to treat cognitive disorders. Trends Pharmacol. Sci. 31, 605–617 (2010).
Tsou, A.Y., Friedman, L.S., Wilson, R.B. & Lynch, D.R. Pharmacotherapy for Friedreich ataxia. CNS Drugs 23, 213–223 (2009).
Day, J.J. & Sweatt, J.D. DNA methylation and memory formation. Nat. Neurosci. 13, 1319–1323 (2010).
Erraji-Benchekroun, L. et al. Molecular aging in human prefrontal cortex is selective and continuous throughout adult life. Biol. Psychiatry 57, 549–558 (2005).
Tang, B. et al. Normal human aging and early-stage schizophrenia share common molecular profiles. Aging Cell 8, 339–342 (2009).
Lu, T. et al. Gene regulation and DNA damage in the ageing human brain. Nature 429, 883–891 (2004).
Szulwach, K.E. et al. 5-hmC–mediated epigenetic dynamics during postnatal neurodevelopment and aging. Nat. Neurosci. 14, 1607–1616 (2011).
Stadler, F. et al. Histone methylation at gene promoters is associated with developmental regulation and region-specific expression of ionotropic and metabotropic glutamate receptors in human brain. J. Neurochem. 94, 324–336 (2005).
Wang, C.M., Tsai, S.N., Yew, T.W., Kwan, Y.W. & Ngai, S.M. Identification of histone methylation multiplicities patterns in the brain of senescence-accelerated prone mouse 8. Biogerontology 11, 87–102 (2010).
Peleg, S. et al. Altered histone acetylation is associated with age-dependent memory impairment in mice. Science 328, 753–756 (2010).
Hargreaves, D.C., Horng, T. & Medzhitov, R. Control of inducible gene expression by signal-dependent transcriptional elongation. Cell 138, 129–145 (2009).
Copray, S., Huynh, J.L., Sher, F., Casaccia-Bonnefil, P. & Boddeke, E. Epigenetic mechanisms facilitating oligodendrocyte development, maturation, and aging. Glia 57, 1579–1587 (2009).
Yankner, B.A., Lu, T. & Loerch, P. The aging brain. Annu. Rev. Pathol. 3, 41–66 (2008).
Ho, L. & Crabtree, G.R. Chromatin remodelling during development. Nature 463, 474–484 (2010).
Fuentes, P., Canovas, J., Berndt, F.A., Noctor, S.C. & Kukuljan, M. CoREST/LSD1 control the development of pyramidal cortical neurons. Cereb. Cortex 22, 1431–1441 (2012).
Hansen, R.S. et al. The DNMT3B DNA methyltransferase gene is mutated in the ICF immunodeficiency syndrome. Proc. Natl. Acad. Sci. USA 96, 14412–14417 (1999).
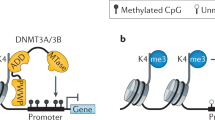
Okano, M., Bell, D.W., Haber, D.A. & Li, E. DNA methyltransferases Dnmt3a and Dnmt3b are essential for de novo methylation and mammalian development. Cell 99, 247–257 (1999).
Jin, B. et al. DNA methyltransferase 3B (DNMT3B) mutations in ICF syndrome lead to altered epigenetic modifications and aberrant expression of genes regulating development, neurogenesis and immune function. Hum. Mol. Genet. 17, 690–709 (2008).
de Greef, J.C. et al. Mutations in ZBTB24 are associated with immunodeficiency, centromeric instability, and facial anomalies syndrome type 2. Am. J. Hum. Genet. 88, 796–804 (2011).
Chouery, E. et al. A novel deletion in ZBTB24 in a Lebanese family with immunodeficiency, centromeric instability and facial anomalies syndrome type 2. Clin. Genet. doi: 10.1111/j.1399-0004.2011.01783.x (2011).
Amir, R.E. et al. Rett syndrome is caused by mutations in X-linked MECP2, encoding methyl-CpG–binding protein-2. Nat. Genet. 23, 185–188 (1999).
Ramocki, M.B. et al. Autism and other neuropsychiatric symptoms are prevalent in individuals with MECP2 duplication syndrome. Ann. Neurol. 66, 771–782 (2009).
Agarwal, N. et al. MeCP2 Rett mutations affect large-scale chromatin organization. Hum. Mol. Genet. 20, 4187–4195. (2011).
Singleton, M.K. et al. MeCP2 is required for global heterochromatic and nucleolar changes during activity-dependent neuronal maturation. Neurobiol. Dis. 43, 190–200 (2011).
Akbarian, S. et al. Expression pattern of the Rett syndrome gene MeCP2 in primate prefrontal cortex. Neurobiol. Dis. 8, 784–791 (2001).
Chen, R.Z., Akbarian, S., Tudor, M. & Jaenisch, R. Deficiency of methyl-CpG–binding protein-2 in CNS neurons results in a Rett-like phenotype in mice. Nat. Genet. 27, 327–331 (2001).
Skene, P.J. et al. Neuronal MeCP2 is expressed at near–histone-octamer levels and globally alters the chromatin state. Mol. Cell 37, 457–468 (2010).
Leonhardt, H., Page, A.W., Weier, H.U. & Bestor, T.H. A targeting sequence directs DNA methyltransferase to sites of DNA replication in mammalian nuclei. Cell 71, 865–873 (1992).
Easwaran, H.P., Schermelleh, L., Leonhardt, H. & Cardoso, M.C. Replication-independent chromatin loading of Dnmt1 during G2 and M phases. EMBO Rep. 5, 1181–1186 (2004).
Fan, G. et al. DNA hypomethylation perturbs the function and survival of CNS neurons in postnatal animals. J. Neurosci. 21, 788–797 (2001).
Bérubé, N.G. et al. The chromatin-remodeling protein ATRX is critical for neuronal survival during corticogenesis. J. Clin. Invest. 115, 258–267 (2005).
Seah, C. et al. Neuronal death resulting from targeted disruption of the Snf2 protein ATRX is mediated by p53. J. Neurosci. 28, 12570–12580 (2008).
Shioda, N. et al. Aberrant calcium/calmodulin–dependent protein kinase II (CaMKII) activity is associated with abnormal dendritic spine morphology in the ATRX mutant mouse brain. J. Neurosci. 31, 346–358 (2011).
Kaufmann, W.E. & Moser, H.W. Dendritic anomalies in disorders associated with mental retardation. Cereb. Cortex 10, 981–991 (2000).
Law, M.J. et al. ATR-X syndrome protein targets tandem repeats and influences allele-specific expression in a size-dependent manner. Cell 143, 367–378 (2010).
Lewis, P.W., Elsaesser, S.J., Noh, K.M., Stadler, S.C. & Allis, C.D. Daxx is an H3.3-specific histone chaperone and cooperates with ATRX in replication-independent chromatin assembly at telomeres. Proc. Natl. Acad. Sci. USA 107, 14075–14080 (2010).
Eustermann, S. et al. Combinatorial readout of histone H3 modifications specifies localization of ATRX to heterochromatin. Nat. Struct. Mol. Biol. 18, 777–782 (2011).
Baumann, C., Viveiros, M.M. & De La Fuente, R. Loss of maternal ATRX results in centromere instability and aneuploidy in the mammalian oocyte and preimplantation embryo. PLoS Genet. 6, e1001137 (2010).
Steffan, J.S. et al. Histone deacetylase inhibitors arrest polyglutamine-dependent neurodegeneration in Drosophila. Nature 413, 739–743 (2001).
Ferrante, R.J. et al. Histone deacetylase inhibition by sodium butyrate chemotherapy ameliorates the neurodegenerative phenotype in Huntington's disease mice. J. Neurosci. 23, 9418–9427 (2003).
Desplats, P. et al. α-synuclein sequesters Dnmt1 from the nucleus: a novel mechanism for epigenetic alterations in Lewy body diseases. J. Biol. Chem. 286, 9031–9037 (2011).
Chestnut, B.A. et al. Epigenetic regulation of motor-neuron cell death through DNA methylation. J. Neurosci. 31, 16619–16636 (2011).
Shock, L.S., Thakkar, P.V., Peterson, E.J., Moran, R.G. & Taylor, S.M. DNA methyltransferase 1, cytosine methylation and cytosine hydroxymethylation in mammalian mitochondria. Proc. Natl. Acad. Sci. USA 108, 3630–3635 (2011).
Nucifora, F.C. Jr. et al. Interference by huntingtin and atrophin-1 with CBP-mediated transcription leading to cellular toxicity. Science 291, 2423–2428 (2001).
Caccamo, A., Maldonado, M.A., Bokov, A.F., Majumder, S. & Oddo, S. CBP gene transfer increases BDNF levels and ameliorates learning and memory deficits in a mouse model of Alzheimer's disease. Proc. Natl. Acad. Sci. USA 107, 22687–22692 (2010).
Wilburn, B. et al. An antisense CAG repeat transcript at JPH3 locus mediates expanded polyglutamine protein toxicity in Huntington's disease–like 2 mice. Neuron 70, 427–440 (2011).
Kazemi-Esfarjani, P. & La Spada, A.R. Déjà vu with a twist: transglutaminases in bioenergetics and transcriptional dysfunction in Huntington's disease. EMBO Mol. Med. 2, 335–337 (2010).
Munsie, L. et al. Mutant huntingtin causes defective actin remodeling during stress: defining a new role for transglutaminase-2 in neurodegenerative disease. Hum. Mol. Genet. 20, 1937–1951 (2011).
McConoughey, S.J. et al. Inhibition of transglutaminase-2 mitigates transcriptional dysregulation in models of Huntington disease. EMBO Mol. Med. 2, 349–370 (2010).
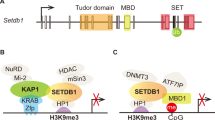
Ryu, H. et al. ESET/SETDB1 gene expression and histone H3 (K9) trimethylation in Huntington's disease. Proc. Natl. Acad. Sci. USA 103, 19176–19181 (2006).
Hu, Y. et al. Transcriptional modulator H2A histone family, member Y (H2AFY) marks Huntington disease activity in man and mouse. Proc. Natl. Acad. Sci. USA 108, 17141–17146 (2011).
Stack, E.C. et al. Modulation of nucleosome dynamics in Huntington's disease. Hum. Mol. Genet. 16, 1164–1175 (2007).
Morris, M.J., Karra, A.S. & Monteggia, L.M. Histone deacetylases govern cellular mechanisms underlying behavioral and synaptic plasticity in the developing and adult brain. Behav. Pharmacol. 21, 409–419 (2010).
Covington, H.E. III et al. Antidepressant actions of histone deacetylase inhibitors. J. Neurosci. 29, 11451–11460 (2009).
Schroeder, F.A., Lin, C.L., Crusio, W.E. & Akbarian, S. Antidepressant-like effects of the histone deacetylase inhibitor, sodium butyrate, in the mouse. Biol. Psychiatry 62, 55–64 (2007).
Zhao, W. et al. Negative regulation of the deacetylase SIRT1 by DBC1. Nature 451, 587–590 (2008).
Dokmanovic, M. & Marks, P.A. Prospects: histone deacetylase inhibitors. J. Cell. Biochem. 96, 293–304 (2005).
Jeong, H. et al. Sirt1 mediates neuroprotection from mutant huntingtin by activation of the TORC1 and CREB transcriptional pathway. Nat. Med. 18, 159–165 (2012).
Libert, S. et al. SIRT1 activates MAO-A in the brain to mediate anxiety and exploratory drive. Cell 147, 1459–1472 (2011).
Rohrer, J.D. et al. Clinical and neuroanatomical signatures of tissue pathology in frontotemporal lobar degeneration. Brain 134, 2565–2581 (2011).
Cenik, B. et al. Suberoylanilide hydroxamic acid (vorinostat) upregulates progranulin transcription: rational therapeutic approach to frontotemporal dementia. J. Biol. Chem. 286, 16101–16108 (2011).
Min, S.W. et al. Acetylation of tau inhibits its degradation and contributes to tauopathy. Neuron 67, 953–966 (2010).
Donmez, G., Wang, D., Cohen, D.E. & Guarente, L. SIRT1 suppresses β-amyloid production by activating the α-secretase gene ADAM10. Cell 142, 320–332 (2010).
Green, K.N. et al. Nicotinamide restores cognition in Alzheimer's disease transgenic mice via a mechanism involving sirtuin inhibition and selective reduction of Thr231-phosphotau. J. Neurosci. 28, 11500–11510 (2008).
Outeiro, T.F. et al. Sirtuin 2 inhibitors rescue α-synuclein–mediated toxicity in models of Parkinson's disease. Science 317, 516–519 (2007).
Hsieh, J., Nakashima, K., Kuwabara, T., Mejia, E. & Gage, F.H. Histone deacetylase inhibition–mediated neuronal differentiation of multipotent adult neural progenitor cells. Proc. Natl. Acad. Sci. USA 101, 16659–16664 (2004).
Pedre, X. et al. Changed histone acetylation patterns in normal-appearing white matter and early multiple sclerosis lesions. J. Neurosci. 31, 3435–3445 (2011).
Liu, J. & Casaccia, P. Epigenetic regulation of oligodendrocyte identity. Trends Neurosci. 33, 193–201 (2010).
Coufal, N.G. et al. Ataxia telangiectasia mutated (ATM) modulates long interspersed element-1 (L1) retrotransposition in human neural stem cells. Proc. Natl. Acad. Sci. USA 108, 20382–20387 (2011).
Rahman, S. et al. The Brd4 extraterminal domain confers transcription activation independent of pTEFb by recruiting multiple proteins, including NSD3. Mol. Cell. Biol. 31, 2641–2652 (2011).
Delmore, J.E. et al. BET bromodomain inhibition as a therapeutic strategy to target c-Myc. Cell 146, 904–917 (2011).
Filippakopoulos, P. et al. Selective inhibition of BET bromodomains. Nature 468, 1067–1073 (2010).
Nicodeme, E. et al. Suppression of inflammation by a synthetic histone mimic. Nature 468, 1119–1123 (2010).
Lein, E.S. et al. Genome-wide atlas of gene expression in the adult mouse brain. Nature 445, 168–176 (2007).
Kubicek, S. et al. Reversal of H3K9me2 by a small-molecule inhibitor for the G9a histone methyltransferase. Mol. Cell 25, 473–481 (2007).
Maze, I. et al. Essential role of the histone methyltransferase G9a in cocaine-induced plasticity. Science 327, 213–216 (2010).
Kelly, T.K., De Carvalho, D.D. & Jones, P.A. Epigenetic modifications as therapeutic targets. Nat. Biotechnol. 28, 1069–1078 (2010).
Levenson, J.M. et al. Evidence that DNA (cytosine-5) methyltransferase regulates synaptic plasticity in the hippocampus. J. Biol. Chem. 281, 15763–15773 (2006).
Han, J. et al. Effect of 5-aza-2-deoxycytidine microinjecting into hippocampus and prelimbic cortex on acquisition and retrieval of cocaine-induced place preference in C57BL/6 mice. Eur. J. Pharmacol. 642, 93–98 (2010).
Miller, C.A. & Sweatt, J.D. Covalent modification of DNA regulates memory formation. Neuron 53, 857–869 (2007).
LaPlant, Q. et al. Dnmt3a regulates emotional behavior and spine plasticity in the nucleus accumbens. Nat. Neurosci. 13, 1137–1143 (2010).
Lubin, F.D., Roth, T.L. & Sweatt, J.D. Epigenetic regulation of BDNF gene transcription in the consolidation of fear memory. J. Neurosci. 28, 10576–10586 (2008).
Miller, C.A. et al. Cortical DNA methylation maintains remote memory. Nat. Neurosci. 13, 664–666 (2010).
Endres, M., Fan, G., Meisel, A., Dirnagl, U. & Jaenisch, R. Effects of cerebral ischemia in mice lacking DNA methyltransferase-1 in postmitotic neurons. Neuroreport 12, 3763–3766 (2001).
Endres, M. et al. DNA methyltransferase contributes to delayed ischemic brain injury. J. Neurosci. 20, 3175–3181 (2000).
Salerno, S. et al. Recent advances in the development of dual topoisomerase I and II inhibitors as anticancer drugs. Curr. Med. Chem. 17, 4270–4290 (2010).
Huang, H.S. et al. Topoisomerase inhibitors unsilence the dormant allele of Ube3a in neurons. Nature 481, 185–189 (2012).
Bressler, J. et al. The SNRPN promoter is not required for genomic imprinting of the Prader-Willi/Angelman domain in mice. Nat. Genet. 28, 232–240 (2001).
Reik, W. Stability and flexibility of epigenetic gene regulation in mammalian development. Nature 447, 425–432 (2007).
Kishino, T., Lalande, M. & Wagstaff, J. UBE3A/E6-AP mutations cause Angelman syndrome. Nat. Genet. 15, 70–73 (1997).
Matsuura, T. et al. De novo truncating mutations in E6-AP ubiquitin-protein ligase gene (UBE3A) in Angelman syndrome. Nat. Genet. 15, 74–77 (1997).
Sutcliffe, J.S. et al. The E6-Ap ubiquitin-protein ligase (UBE3A) gene is localized within a narrowed Angelman syndrome critical region. Genome Res. 7, 368–377 (1997).
Porteus, M.H. & Baltimore, D. Chimeric nucleases stimulate gene targeting in human cells. Science 300, 763 (2003).
Bogdanove, A.J. & Voytas, D.F. TAL effectors: customizable proteins for DNA targeting. Science 333, 1843–1846 (2011).
Miśkiewicz, K. et al. ELP3 controls active zone morphology by acetylating the ELKS family member Bruchpilot. Neuron 72, 776–788 (2011).
Roelfsema, J.H. & Peters, D.J. Rubinstein-Taybi syndrome: clinical and molecular overview. Expert Rev. Mol. Med. 9, 1–16 (2007).
Ehrlich, M. et al. ICF, an immunodeficiency syndrome: DNA methyltransferase 3B involvement, chromosome anomalies and gene dysregulation. Autoimmunity 41, 253–271 (2008).
Michelson, D.J. et al. Evidence report: genetic and metabolic testing on children with global developmental delay: report of the Quality Standards Subcommittee of the American Academy of Neurology and the Practice Committee of the Child Neurology Society. Neurology 77, 1629–1635 (2011).
Adegbola, A., Gao, H., Sommer, S. & Browning, M. A novel mutation in JARID1C/SMCX in a patient with autism spectrum disorder (ASD). Am. J. Med. Genet. A. 146A, 505–511 (2008).
Kleefstra, T. et al. Further clinical and molecular delineation of the 9q subtelomeric deletion syndrome supports a major contribution of EHMT1 haploinsufficiency to the core phenotype. J. Med. Genet. 46, 598–606 (2009).
Kirov, G. et al. De novo CNV analysis implicates specific abnormalities of postsynaptic signalling complexes in the pathogenesis of schizophrenia. Mol. Psychiatry 17, 142–153 (2012).
Verhoeven, W.M., Egger, J.I., Vermeulen, K., van de Warrenburg, B.P. & Kleefstra, T. Kleefstra syndrome in three adult patients: further delineation of the behavioral and neurological phenotype shows aspects of a neurodegenerative course. Am. J. Med. Genet. A. 155, 2409–2415 (2011).
Berdasco, M. et al. Epigenetic inactivation of the Sotos overgrowth syndrome gene histone methyltransferase NSD1 in human neuroblastoma and glioma. Proc. Natl. Acad. Sci. USA 106, 21830–21835 (2009).
Kleine-Kohlbrecher, D. et al. A functional link between the histone demethylase PHF8 and the transcription factor ZNF711 in X-linked mental retardation. Mol. Cell 38, 165–178 (2010).
Fortschegger, K. et al. PHF8 targets histone methylation and RNA polymerase II to activate transcription. Mol. Cell. Biol. 30, 3286–3298 (2010).
Pereira, P.M., Schneider, A., Pannetier, S., Heron, D. & Hanauer, A. Coffin-Lowry syndrome. Eur. J. Hum. Genet. 18, 627–633 (2010).
Percy, A.K. Rett syndrome: exploring the autism link. Arch. Neurol. 68, 985–989 (2011).
Rodríguez-Paredes, M. & Esteller, M. Cancer epigenetics reaches mainstream oncology. Nat. Med. 17, 330–339 (2011).
Li, G. & Reinberg, D. Chromatin higher-order structures and gene regulation. Curr. Opin. Genet. Dev. 21, 175–186 (2011).
Kriaucionis, S. & Heintz, N. The nuclear DNA base 5-hydroxymethylcytosine is present in Purkinje neurons and the brain. Science 324, 929–930 (2009).
Jin, S.G., Wu, X., Li, A.X. & Pfeifer, G.P. Genomic mapping of 5-hydroxymethylcytosine in the human brain. Nucleic Acids Res. 39, 5015–5024 (2011).
Song, C.X. et al. Selective chemical labeling reveals the genome-wide distribution of 5-hydroxymethylcytosine. Nat. Biotechnol. 29, 68–72 (2011).
Maunakea, A.K. et al. Conserved role of intragenic DNA methylation in regulating alternative promoters. Nature 466, 253–257 (2010).
Tan, M. et al. Identification of 67 histone marks and histone lysine crotonylation as a new type of histone modification. Cell 146, 1016–1028 (2011).
Kouzarides, T. Chromatin modifications and their function. Cell 128, 693–705 (2007).
Taverna, S.D., Li, H., Ruthenburg, A.J., Allis, C.D. & Patel, D.J. How chromatin-binding modules interpret histone modifications: lessons from professional pocket pickers. Nat. Struct. Mol. Biol. 14, 1025–1040 (2007).
Zhou, V.W., Goren, A. & Bernstein, B.E. Charting histone modifications and the functional organization of mammalian genomes. Nat. Rev. Genet. 12, 7–18 (2011).
Woodcock, C.L. Chromatin architecture. Curr. Opin. Struct. Biol. 16, 213–220 (2006).
Jin, C. & Felsenfeld, G. Nucleosome stability mediated by histone variants H3.3 and H2A.Z. Genes Dev. 21, 1519–1529 (2007).
Woodcock, C.L., Skoultchi, A.I. & Fan, Y. Role of linker histone in chromatin structure and function: H1 stoichiometry and nucleosome repeat length. Chromosome Res. 14, 17–25 (2006).
Pearson, E.C., Bates, D.L., Prospero, T.D. & Thomas, J.O. Neuronal nuclei and glial nuclei from mammalian cerebral cortex. Nucleosome repeat lengths, DNA contents and H1 contents. Eur. J. Biochem. 144, 353–360 (1984).
Ghosh, R.P., Horowitz-Scherer, R.A., Nikitina, T., Shlyakhtenko, L.S. & Woodcock, C.L. MeCP2 binds cooperatively to its substrate and competes with histone H1 for chromatin binding sites. Mol. Cell. Biol. 30, 4656–4670 (2010).
Henikoff, S. & Shilatifard, A. Histone modification: cause or cog? Trends Genet. 27, 389–396 (2011).
Robison, A.J. & Nestler, E.J. Transcriptional and epigenetic mechanisms of addiction. Nat. Rev. Neurosci. 12, 623–637 (2011).
Day, J.J. & Sweatt, J.D. Epigenetic mechanisms in cognition. Neuron 70, 813–829 (2011).
Guo, J.U., Su, Y., Zhong, C., Ming, G.L. & Song, H. Hydroxylation of 5-methylcytosine by TET1 promotes active DNA demethylation in the adult brain. Cell 145, 423–434 (2011).
Bhutani, N., Burns, D.M. & Blau, H.M. DNA demethylation dynamics. Cell 146, 866–872 (2011).
Copeland, R.A., Solomon, M.E. & Richon, V.M. Protein methyltransferases as a target class for drug discovery. Nat. Rev. Drug Discov. 8, 724–732 (2009).
Rotili, D. & Mai, A. Targeting histone demethylases: a new avenue for the fight against cancer. Genes Canc. 2, 663–679 (2011).
Vermeulen, M. et al. Quantitative-interaction proteomics and genome-wide profiling of epigenetic histone marks and their readers. Cell 142, 967–980 (2010).
Chao, H.T. & Zoghbi, H.Y. MeCP2: only 100% will do. Nat. Neurosci. 15, 176–177 (2012).
Samaco, R.C. et al. Crh and Oprm1 mediate anxiety-related behavior and social approach in a mouse model of MECP2 duplication syndrome. Nat. Genet. 44, 206–211 (2012).
Goffin, D. et al. Rett syndrome mutation MeCP2 T158A disrupts DNA binding, protein stability and ERP responses. Nat. Neurosci. 15, 274–283 (2012).
Tao, J. et al. Phosphorylation of MeCP2 at serine 80 regulates its chromatin association and neurological function. Proc. Natl. Acad. Sci. USA 106, 4882–4887 (2009).
Li, H., Zhong, X., Chau, K.F., Williams, E.C. & Chang, Q. Loss of activity-induced phosphorylation of MeCP2 enhances synaptogenesis, LTP and spatial memory. Nat. Neurosci. 14, 1001–1008 (2011).
Kazazian, H.H. Jr. Mobile elements: drivers of genome evolution. Science 303, 1626–1632 (2004).
Cordaux, R. & Batzer, M.A. The impact of retrotransposons on human genome evolution. Nat. Rev. Genet. 10, 691–703 (2009).
Baillie, J.K. et al. Somatic retrotransposition alters the genetic landscape of the human brain. Nature 479, 534–537 (2011).
Coufal, N.G. et al. L1 retrotransposition in human neural progenitor cells. Nature 460, 1127–1131 (2009).
Singer, T., McConnell, M.J., Marchetto, M.C., Coufal, N.G. & Gage, F.H. LINE-1 retrotransposons: mediators of somatic variation in neuronal genomes? Trends Neurosci. 33, 345–354 (2010).
Muotri, A.R., Zhao, C., Marchetto, M.C. & Gage, F.H. Environmental influence on L1 retrotransposons in the adult hippocampus. Hippocampus 19, 1002–1007 (2009).
Maze, I. et al. Cocaine dynamically regulates heterochromatin and repetitive element unsilencing in nucleus accumbens. Proc. Natl. Acad. Sci. USA 108, 3035–3040 (2011).
Slotkin, R.K. & Martienssen, R. Transposable elements and the epigenetic regulation of the genome. Nat. Rev. Genet. 8, 272–285 (2007).
Muotri, A.R. et al. Somatic mosaicism in neuronal precursor cells mediated by L1 retrotransposition. Nature 435, 903–910 (2005).
Kuwabara, T. et al. Wnt-mediated activation of NeuroD1 and retro-elements during adult neurogenesis. Nat. Neurosci. 12, 1097–1105 (2009).
Muotri, A.R. et al. L1 retrotransposition in neurons is modulated by MeCP2. Nature 468, 443–446 (2010).
Acknowledgements
Work in the authors' laboratory is supported by funds from the US National Institutes of Health (National Institute of Neurological Disorders and Stroke, National Institute of Mental Health, National Institute on Drug Abuse), the US Defense Advanced Research Projects Agency and Autism Speaks.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Rights and permissions
About this article
Cite this article
Jakovcevski, M., Akbarian, S. Epigenetic mechanisms in neurological disease. Nat Med 18, 1194–1204 (2012). https://doi.org/10.1038/nm.2828
Published:
Issue Date:
DOI: https://doi.org/10.1038/nm.2828
This article is cited by
-
WNT signalling control by KDM5C during development affects cognition
Nature (2024)
-
Sequence terminus dependent PCR for site-specific mutation and modification detection
Nature Communications (2023)
-
Neuroepigenetic Changes in DNA Methylation Affecting Diabetes-Induced Cognitive Impairment
Cellular and Molecular Neurobiology (2023)
-
Epigenetic genes and epilepsy — emerging mechanisms and clinical applications
Nature Reviews Neurology (2022)
-
Birth related parameters are important contributors in autism spectrum disorders
Scientific Reports (2022)