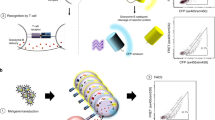
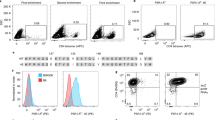
Abstract
Cytotoxic CD8+ T cells recognize the antigenic peptides presented by class I major histocompatibility complex (MHC) molecules. These T cells have key roles in infectious diseases, autoimmunity and tumor immunology, but there is currently no unbiased method for the reliable identification of their target antigens. This is because of the low affinities of antigen-specific T cell receptors (TCR) to their target MHC-peptide complexes, the polyspecificity of these TCRs and the requirement that these TCRs recognize protein antigens that have been processed by antigen-presenting cells (APCs). Here we describe a technology for the unbiased identification of the antigenic peptides presented by MHC class I molecules. The technology uses plasmid-encoded combinatorial peptide libraries and a single-cell detection system. We validated this approach using a well-characterized influenza-virus–specific TCR, MHC and peptide combination. Single APCs carrying antigenic peptides can be detected among several million APCs that carry irrelevant peptides. The identified peptide sequences showed a converging pattern of mimotopes that revealed the parent influenza antigen. This technique should be generally applicable to the identification of disease-relevant T cell antigens.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
References
Appay, V., Douek, D.C. & Price, D.A. CD8+ T cell efficacy in vaccination and disease. Nat. Med. 14, 623–628 (2008).
Kaufmann, S.H. The contribution of immunology to the rational design of novel antibacterial vaccines. Nat. Rev. Microbiol. 5, 491–504 (2007).
Schreiber, R.D., Old, L.J. & Smyth, M.J. Cancer immunoediting: integrating immunity's roles in cancer suppression and promotion. Science 331, 1565–1570 (2011).
Hanahan, D. & Weinberg, R.A. Hallmarks of cancer: the next generation. Cell 144, 646–674 (2011).
Walter, U. & Santamaria, P. CD8+ T cells in autoimmunity. Curr. Opin. Immunol. 17, 624–631 (2005).
Goverman, J. Autoimmune T cell responses in the central nervous system. Nat. Rev. Immunol. 9, 393–407 (2009).
Friese, M.A. & Fugger, L. Pathogenic CD8+ T cells in multiple sclerosis. Ann. Neurol. 66, 132–141 (2009).
Seitz, S. et al. Reconstitution of paired T cell receptor α- and β-chains from microdissected single cells of human inflammatory tissues. Proc. Natl. Acad. Sci. USA 103, 12057–12062 (2006).
Wucherpfennig, K.W. et al. Polyspecificity of T cell and B cell receptor recognition. Semin. Immunol. 19, 216–224 (2007).
Nino-Vasquez, J.J. et al. A powerful combination: the use of positional scanning libraries and biometrical analysis to identify cross-reactive T cell epitopes. Mol. Immunol. 40, 1063–1074 (2004).
Zhao, Y. et al. Combinatorial peptide libraries and biometric score matrices permit the quantitative analysis of specific and degenerate interactions between clonotypic TCR and MHC peptide ligands. J. Immunol. 167, 2130–2141 (2001).
Hemmer, B. et al. Identification of candidate T-cell epitopes and molecular mimics in chronic Lyme disease. Nat. Med. 5, 1375–1382 (1999).
Hammer, G.E., Kanaseki, T. & Shastri, N. The final touches make perfect the peptide-MHC class I repertoire. Immunity 26, 397–406 (2007).
Vyas, J.M., Van der Veen, A.G. & Ploegh, H.L. The known unknowns of antigen processing and presentation. Nat. Rev. Immunol. 8, 607–618 (2008).
Martinon, F., Mayor, A. & Tschopp, J. The inflammasomes: guardians of the body. Annu. Rev. Immunol. 27, 229–265 (2009).
van der Bruggen, P. et al. A gene encoding an antigen recognized by cytolytic T lymphocytes on a human melanoma. Science 254, 1643–1647 (1991).
Wong, F.S. et al. Identification of an MHC class I–restricted autoantigen in type I diabetes by screening an organ-specific cDNA library. Nat. Med. 5, 1026–1031 (1999).
Smith, E.S. et al. Lethality-based selection of recombinant genes in mammalian cells: application to identifying tumor antigens. Nat. Med. 7, 967–972 (2001).
Boon, T., Coulie, P.G., Van den Eynde, B.J. & Van der Bruggen, P. Human T cell responses against melanoma. Annu. Rev. Immunol. 24, 175–208 (2006).
Crawford, F. et al. Use of baculovirus MHC/peptide display libraries to characterize T-cell receptor ligands. Immunol. Rev. 210, 156–170 (2006).
Karttunen, J. & Shastri, N. Measurements of ligand induced activation in single viable T cells using lacZ reporter gene. Proc. Natl. Acad. Sci. USA 88, 3972–3976 (1991).
Fiering, S. et al. Single cell assay of a transcription factor reveals a threshold in transcription activated by signals emanating from the T-cell antigen receptor. Genes Dev. 4, 1823–1834 (1990).
Gotch, F., Rothbard, J., Howland, K., Townsend, A. & McMichael, A. Cytotoxic lymphocytes-T recognize a fragment of influenza-virus matrix protein in association with HLA-A2. Nature 326, 881–882 (1987).
Stewart-Jones, G.B.E., McMichael, A.J., Bell, J.I., Stuart, D.I. & Jones, E.Y. A structural basis for immunodominant human T cell receptor recognition. Nat. Immunol. 4, 657–663 (2003).
Ishizuka, J. et al. The structural dynamics and energetics of an immunodominant T cell receptor are programmed by its Vβ domain. Immunity 28, 171–182 (2008).
Blank, U., Boitel, B., Mège, D., Ermonval, M. & Acuto, O. Analysis of tetanus toxin peptide/DR recognition by human T cell receptors reconstituted into a murine T cell hybridoma. Eur. J. Immunol. 23, 3057–3065 (1993).
Rammensee, H., Bachmann, J., Emmerich, N.P., Bachor, O.A. & Stevanovic, S. SYFPEITHI: database for MHC ligands and peptide motifs. Immunogenetics 50, 213–219 (1999).
Kawakami, N. et al. Live imaging of effector cell trafficking and autoantigen recognition within the unfolding autoimmune encephalomyelitis lesion. J. Exp. Med. 201, 1805–1814 (2005).
Venturi, V., Price, D.A., Douek, D.C. & Davenport, M.P. The molecular basis for public T-cell responses? Nat. Rev. Immunol. 8, 231–238 (2008).
Bansal, A. et al. CD8 T cell response and evolutionary pressure to HIV-1 cryptic epitopes derived from antisense transcription. J. Exp. Med. 89, 51–59 (2010).
Bendle, G.M. et al. Lethal graft-versus-host disease in mouse models of T cell receptor gene therapy. Nat. Med. 16, 565–570 (2010).
Lehner, P.J. et al. Human HLA-A0201-restricted cytotoxic T lymphocyte recognition of influenza A is dominated by T cells bearing the Vβ17 gene segment. J. Exp. Med. 181, 79–91 (1995).
Heim, R., Cubitt, A.B. & Tsien, R.Y. Improved green fluorescence. Nature 373, 663–664 (1995).
Bettinotti, M.P. et al. New HLA-A, -B, and -C locus–specific primers for PCR amplification from cDNA: application in clinical immunology. J. Immunol. Methods 279, 143–148 (2003).
Pircher, H. et al. T cell tolerance to Mlsa encoded antigens in T cell receptor Vβ8.1 chain transgenic mice. EMBO J. 8, 719–727 (1989).
Gluzman, Y. SV40-transformed simian cells support the replication of early SV40 mutants. Cell 23, 175–182 (1981).
Acknowledgements
We thank E. Meinl and W.E.F. Klinkert for comments on the manuscript, D. Hackl and M. Ackmann for help with synthesizing the plasmid pcDNA-NFAT-sGFP, I. Bartholomäus and M. Mues for advice regarding fluorescence microscopy, I. Eiglmeier for expert technical assistance and K. Ogston for editing the manuscript. This work was supported by the Deutsche Forschungsgemeinschaft (grant SFB 571 A1).
Author information
Authors and Affiliations
Contributions
K.S. designed and performed most of the experiments and contributed to the writing of the manuscript, J.M. contributed to cloning experiments, N.K. contributed to microscopy experiments, H.W. and R.H. supervised and supported the study and contributed to the writing of the manuscript, and K.D. conceived of, designed and supervised the research and wrote the manuscript.
Corresponding author
Ethics declarations
Competing interests
The Ludwig-Maximilians-University, Munich, Germany, has filed European and International patent applications describing the technology used in this study (K.D., K.S. and R.H.).
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–4, Supplementary Table 1 and Supplementary Methods (PDF 6560 kb)
Supplementary Video 1
Activation of a 58-JM22-CD8-sGFP cell. Time-lapse fluorescence microscopy of a coculture of 58-JM22-CD8-sGFP cells and COS-7 cells. (AVI 1060 kb)
Supplementary Video 2
Isolation of an antigen-expressing cell. We show the isolation of an activated green fluorescent 58-JM22-CD8-sGFP cell together with a subjacent antigen-expressing COS-7 cell. (AVI 1171 kb)
Rights and permissions
About this article
Cite this article
Siewert, K., Malotka, J., Kawakami, N. et al. Unbiased identification of target antigens of CD8+ T cells with combinatorial libraries coding for short peptides. Nat Med 18, 824–828 (2012). https://doi.org/10.1038/nm.2720
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nm.2720
This article is cited by
-
Rapid TCR:Epitope Ranker (RAPTER): a primary human T cell reactivity screening assay pairing epitope and TCR at single cell resolution
Scientific Reports (2023)
-
Fundamental mechanistic insights from rare but paradigmatic neuroimmunological diseases
Nature Reviews Neurology (2021)
-
Deciphering CD4+ T cell specificity using novel MHC–TCR chimeric receptors
Nature Immunology (2019)
-
Immunopathology of multiple sclerosis
Nature Reviews Immunology (2015)
-
Toward the Clonotype Analysis of Alopecia Areata-Specific, Intralesional Human CD8+ T Lymphocytes
Journal of Investigative Dermatology Symposium Proceedings (2015)