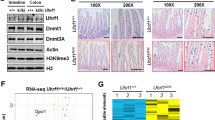
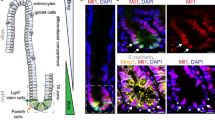
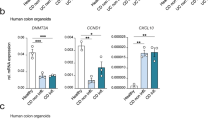
Abstract
Although aberrant DNA methylation is considered to be one of the key ways by which tumor-suppressor and DNA-repair genes are silenced during tumor initiation and progression, the mechanisms underlying DNA methylation alterations in cancer remain unclear. Here we show that prostaglandin E2 (PGE2) silences certain tumor-suppressor and DNA-repair genes through DNA methylation to promote tumor growth. These findings uncover a previously unrecognized role for PGE2 in the promotion of tumor progression.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout


Similar content being viewed by others
References
Wang, D. & DuBois, R.N. Oncogene 29, 781–788 (2010).
Wang, D. & DuBois, R.N. Nat. Rev. Cancer 10, 181–193 (2010).
Baba, Y. et al. Cancer Epidemiol. Biomarkers Prev. 19, 822–831 (2010).
Slattery, M.L. et al. Int. J. Cancer 120, 656–663 (2007).
Kim, M.S., Lee, J. & Sidransky, D. Cancer Metastasis Rev. 29, 181–206 (2010).
Kim, Y.H. et al. Genes Chromosom. Cancer 45, 781–789 (2006).
Wang, D. et al. Cancer Res. 68, 6468–6476 (2008).
De Marzo, A.M. et al. Cancer Res. 59, 3855–3860 (1999).
Schmidt, W.M. et al. Mol. Carcinog. 46, 766–772 (2007).
Nosho, K. et al. Clin. Cancer Res. 15, 3663–3671 (2009).
Ibrahim, A.E. et al. Gut 60, 499–508 (2011).
Kanai, Y., Ushijima, S., Kondo, Y., Nakanishi, Y. & Hirohashi, S. Int. J. Cancer 91, 205–212 (2001).
Steine, E.J. et al. J. Clin. Invest. 121, 1748–1752 (2011).
Lin, H. et al. Mol. Cell Biol. 26, 2976–2983 (2006).
Linhart, H.G. et al. Genes Dev. 21, 3110–3122 (2007).
Shteper, P.J. et al. Oncogene 22, 7737–7749 (2003).
Pakneshan, P., Szyf, M., Farias-Eisner, R. & Rabbani, S.A. J. Biol. Chem. 279, 31735–31744 (2004).
Hamm, C.A. et al. PLoS ONE 4, e8340 (2009).
Ogino, S. et al. J. Natl. Cancer Inst. 100, 1734–1738 (2008).
Ahn, J.B. et al. Cancer 117, 1847–1854 (2011).
Acknowledgements
We thank D. Menter and P. Yang for their suggestions and help in the measurement of PGE2. This work is supported, in part, by the US National Institutes of Health MERIT award grants R37 DK47297, RO1 DK62112, NCI P01 CA77839 and CPRIT RP100960. We also thank the National Colorectal Cancer Research Alliance (NCCRA) for its generous support (R.N.D.) and a cancer prevention fellowship (D.X.) supported by the National Cancer Institute grant R25T CA57730 (principal investigator, S. Chang).
Author information
Authors and Affiliations
Contributions
R.N.D., D.W. and D.X. designed this research project. D.X. performed most of the experiments. S.-H.K. contributed to establish the DNMT1 and DNMT3B knockdown stable cell lines, and H.K. conducted the DNA methylation analysis for the human tissues samples. D.X. and D.W. conducted the data analyses. D.W. wrote the manuscript with help from D.X. R.N.D. supervised the project.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–5, Supplementary Tables 1 and 2, Supplementary Discussion and Supplementary Methods (PDF 2314 kb)
Rights and permissions
About this article
Cite this article
Xia, D., Wang, D., Kim, SH. et al. Prostaglandin E2 promotes intestinal tumor growth via DNA methylation. Nat Med 18, 224–226 (2012). https://doi.org/10.1038/nm.2608
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nm.2608
This article is cited by
-
Reprogramming of arachidonate metabolism confers temozolomide resistance to glioblastoma through enhancing mitochondrial activity in fatty acid oxidation
Journal of Biomedical Science (2022)
-
Targeting cancer-promoting inflammation — have anti-inflammatory therapies come of age?
Nature Reviews Clinical Oncology (2021)
-
Metabolism pathways of arachidonic acids: mechanisms and potential therapeutic targets
Signal Transduction and Targeted Therapy (2021)
-
Protein phosphatase 2A inactivation induces microsatellite instability, neoantigen production and immune response
Nature Communications (2021)
-
PGE2-JNK signaling axis non-canonically promotes Gli activation by protecting Gli2 from ubiquitin-proteasomal degradation
Cell Death & Disease (2021)