Abstract
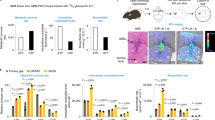
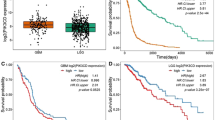
PICT1 (also known as GLTSCR2) is considered a tumor suppressor because it stabilizes phosphatase and tensin homolog (PTEN), but individuals with oligodendrogliomas lacking chromosome 19q13, where PICT1 is located, have better prognoses than other oligodendroglioma patients. To clarify the function of PICT1, we generated Pict1-deficient mice and embryonic stem (ES) cells. Pict1 is a nucleolar protein essential for embryogenesis and ES cell survival. Even without DNA damage, Pict1 loss led to p53-dependent arrest of cell cycle phase G1 and apoptosis. Pict1-deficient cells accumulated p53, owing to impaired Mdm2 function. Pict1 binds Rpl11, and Rpl11 is released from nucleoli in the absence of Pict1. In Pict1-deficient cells, increased binding of Rpl11 to Mdm2 blocks Mdm2-mediated ubiquitination of p53. In human cancer, individuals whose tumors express less PICT1 have better prognoses. When PICT1 is depleted in tumor cells with intact P53 signaling, the cells grow more slowly and accumulate P53. Thus, PICT1 is a potent regulator of the MDM2-P53 pathway and promotes tumor progression by retaining RPL11 in the nucleolus.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
References
Toledo, F. & Wahl, G.M. Regulating the p53 pathway: in vitro hypotheses, in vivo veritas. Nat. Rev. Cancer 6, 909–923 (2006).
Kubbutat, M.H., Jones, S.N. & Vousden, K.H. Regulation of p53 stability by Mdm2. Nature 387, 299–303 (1997).
Haupt, Y., Maya, R., Kazaz, A. & Oren, M. Mdm2 promotes the rapid degradation of p53. Nature 387, 296–299 (1997).
Kruse, J.P. & Gu, W. Modes of p53 regulation. Cell 137, 609–622 (2009).
Zhang, Y., Xiong, Y. & Yarbrough, W.G. ARF promotes MDM2 degradation and stabilizes p53: ARF-INK4a locus deletion impairs both the Rb and p53 tumor suppression pathways. Cell 92, 725–734 (1998).
Pomerantz, J. et al. The Ink4a tumor suppressor gene product, p19Arf, interacts with MDM2 and neutralizes MDM2′s inhibition of p53. Cell 92, 713–723 (1998).
Lindström, M.S., Deisenroth, C. & Zhang, Y. Putting a finger on growth surveillance: insight into MDM2 zinc finger-ribosomal protein interactions. Cell Cycle 6, 434–437 (2007).
Lohrum, M.A., Ludwig, R.L., Kubbutat, M.H., Hanlon, M. & Vousden, K.H. Regulation of HDM2 activity by the ribosomal protein L11. Cancer Cell 3, 577–587 (2003).
Zhang, Y. et al. Ribosomal protein L11 negatively regulates oncoprotein MDM2 and mediates a p53-dependent ribosomal-stress checkpoint pathway. Mol. Cell. Biol. 23, 8902–8912 (2003).
Dai, M.S. & Lu, H. Inhibition of MDM2-mediated p53 ubiquitination and degradation by ribosomal protein L5. J. Biol. Chem. 279, 44475–44482 (2004).
Jin, A., Itahana, K., O'Keefe, K. & Zhang, Y. Inhibition of HDM2 and activation of p53 by ribosomal protein L23. Mol. Cell. Biol. 24, 7669–7680 (2004).
Zhu, Y. et al. Ribosomal protein S7 is both a regulator and a substrate of MDM2. Mol. Cell 35, 316–326 (2009).
Fumagalli, S. et al. Absence of nucleolar disruption after impairment of 40S ribosome biogenesis reveals an rpL11-translation-dependent mechanism of p53 induction. Nat. Cell Biol. 11, 501–508 (2009).
Zhang, Y. & Lu, H. Signaling to p53: ribosomal proteins find their way. Cancer Cell 16, 369–377 (2009).
Warner, J.R. & McIntosh, K.B. How common are extraribosomal functions of ribosomal proteins? Mol. Cell 34, 3–11 (2009).
Chen, D. et al. Ribosomal protein S7 as a novel modulator of p53–MDM2 interaction: binding to MDM2, stabilization of p53 protein, and activation of p53 function. Oncogene 26, 5029–5037 (2007).
Bhat, K.P., Itahana, K., Jin, A. & Zhang, Y. Essential role of ribosomal protein L11 in mediating growth inhibition-induced p53 activation. EMBO J. 23, 2402–2412 (2004).
Rubbi, C.P. & Milner, J. Disruption of the nucleolus mediates stabilization of p53 in response to DNA damage and other stresses. EMBO J. 22, 6068–6077 (2003).
Sun, X.X., Dai, M.S. & Lu, H. Mycophenolic acid activation of p53 requires ribosomal proteins L5 and L11. J. Biol. Chem. 283, 12387–12392 (2008).
Sulic, S. et al. Inactivation of S6 ribosomal protein gene in T lymphocytes activates a p53-dependent checkpoint response. Genes Dev. 19, 3070–3082 (2005).
Takagi, M., Absalon, M.J., McLure, K.G. & Kastan, M.B. Regulation of p53 translation and induction after DNA damage by ribosomal protein L26 and nucleolin. Cell 123, 49–63 (2005).
Yadavilli, S. et al. Ribosomal protein S3: a multi-functional protein that interacts with both p53 and MDM2 through its KH domain. DNA Repair (Amst.) 8, 1215–1224 (2009).
Smith, J.S. et al. A transcript map of the chromosome 19q-arm glioma tumor suppressor region. Genomics 64, 44–50 (2000).
Kim, Y.J. et al. Suppression of putative tumour suppressor gene GLTSCR2 expression in human glioblastomas. J. Pathol. 216, 218–224 (2008).
Nakagawa, Y. et al. Chromosomal imbalances in malignant peripheral nerve sheath tumor detected by metaphase and microarray comparative genomic hybridization. Oncol. Rep. 15, 297–303 (2006).
Merritt, M.A. et al. Expression profiling identifies genes involved in neoplastic transformation of serous ovarian cancer. BMC Cancer 9, 378 (2009).
Yim, J.H. et al. The putative tumor suppressor gene GLTSCR2 induces PTEN-modulated cell death. Cell Death Differ. 14, 1872–1879 (2007).
Okahara, F., Ikawa, H., Kanaho, Y. & Maehama, T. Regulation of PTEN phosphorylation and stability by a tumor suppressor candidate protein. J. Biol. Chem. 279, 45300–45303 (2004).
Okahara, F. et al. Critical role of PICT-1, a tumor suppressor candidate, in phosphatidylinositol 3,4,5-trisphosphate signals and tumorigenic transformation. Mol. Biol. Cell 17, 4888–4895 (2006).
Cairncross, J.G. et al. Specific genetic predictors of chemotherapeutic response and survival in patients with anaplastic oligodendrogliomas. J. Natl. Cancer Inst. 90, 1473–1479 (1998).
Smith, J.S. et al. Localization of common deletion regions on 1p and 19q in human gliomas and their association with histological subtype. Oncogene 18, 4144–4152 (1999).
Mariani, L. et al. Loss of heterozygosity 1p36 and 19q13 is a prognostic factor for overall survival in patients with diffuse WHO grade 2 gliomas treated without chemotherapy. J. Clin. Oncol. 24, 4758–4763 (2006).
von Deimling, A. et al. Loci associated with malignant progression in astrocytomas: a candidate on chromosome 19q. Cancer Res. 54, 1397–1401 (1994).
Smith, J.S. et al. Alterations of chromosome arms 1p and 19q as predictors of survival in oligodendrogliomas, astrocytomas, and mixed oligoastrocytomas. J. Clin. Oncol. 18, 636–645 (2000).
Miyazaki, M. et al. Thymocyte proliferation induced by pre–T cell receptor signaling is maintained through polycomb gene product Bmi-1–mediated Cdkn2a repression. Immunity 28, 231–245 (2008).
Kamijo, T. et al. Functional and physical interactions of the ARF tumor suppressor with p53 and Mdm2. Proc. Natl. Acad. Sci. USA 95, 8292–8297 (1998).
Solozobova, V. & Blattner, C. Regulation of p53 in embryonic stem cells. Exp. Cell Res. 316, 2434–2446 (2010).
Zhao, X. et al. The HECT-domain ubiquitin ligase Huwe1 controls neural differentiation and proliferation by destabilizing the N-Myc oncoprotein. Nat. Cell Biol. 10, 643–653 (2008).
Barak, Y., Juven, T., Haffner, R. & Oren, M. mdm2 expression is induced by wild type p53 activity. EMBO J. 12, 461–468 (1993).
Kurki, S. et al. Nucleolar protein NPM interacts with HDM2 and protects tumor suppressor protein p53 from HDM2-mediated degradation. Cancer Cell 5, 465–475 (2004).
Dai, M.S., Sun, X.X. & Lu, H. Aberrant expression of nucleostemin activates p53 and induces cell cycle arrest via inhibition of MDM2. Mol. Cell. Biol. 28, 4365–4376 (2008).
Saxena, A., Rorie, C.J., Dimitrova, D., Daniely, Y. & Borowiec, J.A. Nucleolin inhibits Hdm2 by multiple pathways leading to p53 stabilization. Oncogene 25, 7274–7288 (2006).
Russo, A. et al. The TP53 colorectal cancer international collaborative study on the prognostic and predictive significance of p53 mutation: influence of tumor site, type of mutation, and adjuvant treatment. J. Clin. Oncol. 23, 7518–7528 (2005).
Egashira, A. et al. p53 gene mutations in esophageal squamous cell carcinoma and their relevance to etiology and pathogenesis: results in Japan and comparisons with other countries. Cancer Sci. 98, 1152–1156 (2007).
Kim, J.Y., Kim, H.S., Lee, S. & Park, J.H. The expression of GLTSCR2, a candidate tumor suppressor, is reduced in seborrheic keratosis compared to normal skin. Pathol. Res. Pract. 206, 295–299 (2010).
Okamoto, Y. et al. Population-based study on incidence, survival rates, and genetic alterations of low-grade diffuse astrocytomas and oligodendrogliomas. Acta Neuropathol. 108, 49–56 (2004).
Andersen, J.S. et al. Nucleolar proteome dynamics. Nature 433, 77–83 (2005).
Era, T. & Witte, O.N. Regulated expression of P210 Bcr-Abl during embryonic stem cell differentiation stimulates multipotential progenitor expansion and myeloid cell fate. Proc. Natl. Acad. Sci. USA 97, 1737–1742 (2000).
Kitajima, K., Masuhara, M., Era, T., Enver, T. & Nakano, T. GATA-2 and GATA-2/ER display opposing activities in the development and differentiation of blood progenitors. EMBO J. 21, 3060–3069 (2002).
Okahara, F. et al. Production of research-grade antibody by in vivo electroporation of DNA-encoding target protein. Anal. Biochem. 336, 138–140 (2005).
Inoue-Narita, T. et al. Pten deficiency in melanocytes results in resistance to hair graying and susceptibility to carcinogen-induced melanomagenesis. Cancer Res. 68, 5760–5768 (2008).
Furukawa, M., Zhang, Y., McCarville, J., Ohta, T. & Xiong, Y. The CUL1 C-terminal sequence and ROC1 are required for efficient nuclear accumulation, NEDD8 modification, and ubiquitin ligase activity of CUL1. Mol. Cell. Biol. 20, 8185–8197 (2000).
Feng, L., Lin, T., Uranishi, H., Gu, W. & Xu, Y. Functional analysis of the roles of posttranslational modifications at the p53 C terminus in regulating p53 stability and activity. Mol. Cell. Biol. 25, 5389–5395 (2005).
Acknowledgements
We are grateful to T. Noda, S. Kuroda, H. Kishimoto, M. Natsui, H. Takahashi, H. Tashiro, N. Yasui, M. Suzuki, S. Suzuki, T. Shono and T. Sasaki for expert technical support and helpful discussions. We also thank H. Miyoshi (RIKEN BioResource Center, Tsukuba, Japan) for providing lentivirus vector plasmid DNA. This work was supported by grants from the Ministry of Education, Culture, Sports and Technology of Japan (MEXT), Takeda Medical Foundation, Naito Foundation, Ono Medical Research Foundation, Yasuda Medical Foundation, and Astellas Foundation for Research on Metabolic Disorders. K.M. and M.M. are supported by the Core Research for Evolutionary Science and Technology program of the Japanese Science and Technology Agency. T.Y., T.S., M.M. and A.S. are supported by the Global Centers of Excellence Program of MEXT.
Author information
Authors and Affiliations
Contributions
M.S. carried out the initial generation and analyses of Pict1flox mice and Pict1 ES cells. K.K. carried out subsequent major biochemical and biological experiments, and M.N. carried out mouse work. K.M., R.K. and M.M. carried out the human cancer tissue analyses. K.H. generated Pict1−/− mice. B.I. assisted with confocal microscopy. J.W., Y.K. and Y.R.Y. assisted with the introduction of shRNA into human cancer cell lines. H.H. assisted with the protein binding assays. Y.H. carried out mouse analyses. T.Y., T.K., Y. Zhang, Y. Zhu, C.P. and T.W.M. provided key materials. T.M., K.M. and A.S. conceived of the project, and M.S., K.K., K.M, T.M., M.M. and A.S. designed the experiments. M.S., K.K., M.N., K.M., R.K., T.Y., T.K., Y. Zhang, C.P., T.N., T.W.M., T.S., T.M., M.M. and A.S. discussed the hypothesis and interpreted the data. A.S. coordinated and directed the project and wrote the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–8 and Supplementary Methods (PDF 689 kb)
Rights and permissions
About this article
Cite this article
Sasaki, M., Kawahara, K., Nishio, M. et al. Regulation of the MDM2-P53 pathway and tumor growth by PICT1 via nucleolar RPL11. Nat Med 17, 944–951 (2011). https://doi.org/10.1038/nm.2392
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nm.2392
This article is cited by
-
Changes in calpain-2 expression during glioblastoma progression predisposes tumor cells to temozolomide resistance by minimizing DNA damage and p53-dependent apoptosis
Cancer Cell International (2023)
-
High filamin-C expression predicts enhanced invasiveness and poor outcome in glioblastoma multiforme
British Journal of Cancer (2019)
-
Nucleolus as an emerging hub in maintenance of genome stability and cancer pathogenesis
Oncogene (2018)
-
Sequencing of prostate cancers identifies new cancer genes, routes of progression and drug targets
Nature Genetics (2018)
-
Preribosomes escaping from the nucleus are caught during translation by cytoplasmic quality control
Nature Structural & Molecular Biology (2017)