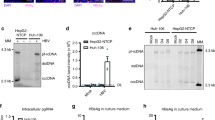
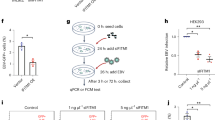
Abstract
Hepatitis C virus (HCV) is a major cause of liver disease, but therapeutic options are limited and there are no prevention strategies. Viral entry is the first step of infection and requires the cooperative interaction of several host cell factors. Using a functional RNAi kinase screen, we identified epidermal growth factor receptor and ephrin receptor A2 as host cofactors for HCV entry. Blocking receptor kinase activity by approved inhibitors broadly impaired infection by all major HCV genotypes and viral escape variants in cell culture and in a human liver chimeric mouse model in vivo. The identified receptor tyrosine kinases (RTKs) mediate HCV entry by regulating CD81–claudin-1 co-receptor associations and viral glycoprotein–dependent membrane fusion. These results identify RTKs as previously unknown HCV entry cofactors and show that tyrosine kinase inhibitors have substantial antiviral activity. Inhibition of RTK function may constitute a new approach for prevention and treatment of HCV infection.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
References
Tai, A.W. & Chung, R.T. Treatment failure in hepatitis C: mechanisms of non-response. J. Hepatol. 50, 412–420 (2009).
Hézode, C. et al. Telaprevir and peginterferon with or without ribavirin for chronic HCV infection. N. Engl. J. Med. 360, 1839–1850 (2009).
Timpe, J.M. et al. Hepatitis C virus cell-cell transmission in hepatoma cells in the presence of neutralizing antibodies. Hepatology 47, 17–24 (2008).
Zeisel, M.B., Cosset, F.L. & Baumert, T.F. Host neutralizing responses and pathogenesis of hepatitis C virus infection. Hepatology 48, 299–307 (2008).
von Hahn, T. & Rice, C.M. Hepatitis C virus entry. J. Biol. Chem. 283, 3689–3693 (2008).
Barth, H. et al. Viral and cellular determinants of the hepatitis C virus envelope–heparan sulfate interaction. J. Virol. 80, 10579–10590 (2006).
Pileri, P. et al. Binding of hepatitis C virus to CD81. Science 282, 938–941 (1998).
Scarselli, E. et al. The human scavenger receptor class B type I is a novel candidate receptor for the hepatitis C virus. EMBO J. 21, 5017–5025 (2002).
Evans, M.J. et al. Claudin-1 is a hepatitis C virus co-receptor required for a late step in entry. Nature 446, 801–805 (2007).
Ploss, A. et al. Human occludin is a hepatitis C virus entry factor required for infection of mouse cells. Nature 457, 882–886 (2009).
Hidalgo, M. & Bloedow, D. Pharmacokinetics and pharmacodynamics: maximizing the clinical potential of erlotinib (Tarceva). Semin. Oncol. 30, 25–33 (2003).
Shepherd, F.A. et al. Erlotinib in previously treated non–small-cell lung cancer. N. Engl. J. Med. 353, 123–132 (2005).
Li, J. et al. A chemical and phosphoproteomic characterization of dasatinib action in lung cancer. Nat. Chem. Biol. 6, 291–299 (2010).
Fafi-Kremer, S. et al. Viral entry and escape from antibody-mediated neutralization influence hepatitis C virus reinfection in liver transplantation. J. Exp. Med. 207, 2019–2031 (2010).
Mee, C.J. et al. Polarization restricts hepatitis C virus entry into HepG2 hepatoma cells. J. Virol. 83, 6211–6221 (2009).
Schlessinger, J. Ligand-induced, receptor-mediated dimerization and activation of EGF receptor. Cell 110, 669–672 (2002).
Thoresen, G.H. et al. Response to transforming growth factor α (TGFα) and epidermal growth factor (EGF) in hepatocytes: lower EGF receptor affinity of TGFα is associated with more sustained activation of p42/p44 mitogen-activated protein kinase and greater efficacy in stimulation of DNA synthesis. J. Cell. Physiol. 175, 10–18 (1998).
Dreux, M. et al. Receptor complementation and mutagenesis reveal SR-BI as an essential HCV entry factor and functionally imply its intra- and extra-cellular domains. PLoS Pathog. 5, e1000310 (2009).
Krieger, S.E. et al. Inhibition of hepatitis C virus infection by anti–claudin-1 antibodies is mediated by neutralization of E2–CD81–claudin-1 associations. Hepatology 51, 1144–1157 (2010).
Koutsoudakis, G. et al. Characterization of the early steps of hepatitis C virus infection by using luciferase reporter viruses. J. Virol. 80, 5308–5320 (2006).
Zeisel, M.B. et al. Scavenger receptor class B type I is a key host factor for hepatitis C virus infection required for an entry step closely linked to CD81. Hepatology 46, 1722–1731 (2007).
Dreux, M. et al. High density lipoprotein inhibits hepatitis C virus-neutralizing antibodies by stimulating cell entry via activation of the scavenger receptor BI. J. Biol. Chem. 281, 18285–18295 (2006).
Harris, H.J. et al. Claudin association with CD81 defines hepatitis C virus entry. J. Biol. Chem. 285, 21092–21102 (2010).
Harris, H.J. et al. CD81 and claudin 1 coreceptor association: role in hepatitis C virus entry. J. Virol. 82, 5007–5020 (2008).
Lavillette, D. et al. Characterization of fusion determinants points to the involvement of three discrete regions of both E1 and E2 glycoproteins in the membrane fusion process of hepatitis C virus. J. Virol. 81, 8752–8765 (2007).
Witteveldt, J. et al. CD81 is dispensable for hepatitis C virus cell-to-cell transmission in hepatoma cells. J. Gen. Virol. 90, 48–58 (2009).
Hiraga, N. et al. Absence of viral interference and different susceptibility to interferon between hepatitis B virus and hepatitis C virus in human hepatocyte chimeric mice. J. Hepatol. 51, 1046–1054 (2009).
Kamiya, N. et al. Practical evaluation of a mouse with chimeric human liver model for hepatitis C virus infection using an NS3–4A protease inhibitor. J. Gen. Virol. 91, 1668–1677 (2010).
Meuleman, P. et al. Morphological and biochemical characterization of a human liver in a uPA-SCID mouse chimera. Hepatology 41, 847–856 (2005).
Higgins, B. et al. Antitumor activity of erlotinib (OSI-774, Tarceva) alone or in combination in human non–small cell lung cancer tumor xenograft models. Anticancer Drugs 15, 503–512 (2004).
Schneider, M.R. & Wolf, E. The epidermal growth factor receptor ligands at a glance. J. Cell. Physiol. 218, 460–466 (2009).
Lackmann, M. & Boyd, A.W. Eph, a protein family coming of age: more confusion, insight, or complexity? Sci. Signal. 1, re2 (2008).
Singh, A.B. & Harris, R.C. Epidermal growth factor receptor activation differentially regulates claudin expression and enhances transepithelial resistance in Madin-Darby canine kidney cells. J. Biol. Chem. 279, 3543–3552 (2004).
Flores-Benitez, D. et al. Control of tight junctional sealing: roles of epidermal growth factor and prostaglandin E2. Am. J. Physiol. Cell Physiol. 297, C611–C620 (2009).
Blanchard, E. et al. Hepatitis C virus entry depends on clathrin-mediated endocytosis. J. Virol. 80, 6964–6972 (2006).
Reiter, J.L. & Maihle, N.J. A 1.8 kb alternative transcript from the human epidermal growth factor receptor gene encodes a truncated form of the receptor. Nucleic Acids Res. 24, 4050–4056 (1996).
McCole, D.F., Keely, S.J., Coffey, R.J. & Barrett, K.E. Transactivation of the epidermal growth factor receptor in colonic epithelial cells by carbachol requires extracellular release of transforming growth factor-α. J. Biol. Chem. 277, 42603–42612 (2002).
Miao, H. et al. EphA2 mediates ligand-dependent inhibition and ligand-independent promotion of cell migration and invasion via a reciprocal regulatory loop with Akt. Cancer Cell 16, 9–20 (2009).
Pestka, J.M. et al. Rapid induction of virus-neutralizing antibodies and viral clearance in a single-source outbreak of hepatitis C. Proc. Natl. Acad. Sci. USA 104, 6025–6030 (2007).
Rothenberg, S.M. et al. Modeling oncogene addiction using RNA interference. Proc. Natl. Acad. Sci. USA 105, 12480–12484 (2008).
Bartosch, B., Dubuisson, J. & Cosset, F.L. Infectious hepatitis C virus pseudo-particles containing functional E1–E2 envelope protein complexes. J. Exp. Med. 197, 633–642 (2003).
Pietschmann, T. et al. Construction and characterization of infectious intragenotypic and intergenotypic hepatitis C virus chimeras. Proc. Natl. Acad. Sci. USA 103, 7408–7413 (2006).
Kato, T. et al. Efficient replication of the genotype 2a hepatitis C virus subgenomic replicon. Gastroenterology 125, 1808–1817 (2003).
Lavillette, D. et al. Characterization of host-range and cell entry properties of the major genotypes and subtypes of hepatitis C virus. Hepatology 41, 265–274 (2005).
Frecha, C. et al. Efficient and stable transduction of resting B lymphocytes and primary chronic lymphocyte leukemia cells using measles virus GP displaying lentiviral vectors. Blood 114, 3173–3180 (2009).
Sandrin, V. et al. Lentiviral vectors pseudotyped with a modified RD114 envelope glycoprotein show increased stability in sera and augmented transduction of primary lymphocytes and CD34+ cells derived from human and nonhuman primates. Blood 100, 823–832 (2002).
Fofana, I. et al. Monoclonal anti–claudin 1 antibodies prevent hepatitis C virus infection of primary human hepatocytes. Gastroenterology 139, 953–964, 964.e1–e4 (2010).
Meunier, J.C. et al. Isolation and characterization of broadly neutralizing human monoclonal antibodies to the e1 glycoprotein of hepatitis C virus. J. Virol. 82, 966–973 (2008).
Lohmann, V. et al. Replication of subgenomic hepatitis C virus RNAs in a hepatoma cell line. Science 285, 110–113 (1999).
van den Heuvel, S. & Harlow, E. Distinct roles for cyclin-dependent kinases in cell cycle control. Science 262, 2050–2054 (1993).
Wang, Y. et al. Negative regulation of EphA2 receptor by Cbl. Biochem. Biophys. Res. Commun. 296, 214–220 (2002).
Chahbouni, A., den Burger, J.C., Vos, R.M., Sinjewel, A. & Wilhelm, A.J. Simultaneous quantification of erlotinib, gefitinib and imatinib in human plasma by liquid chromatography tandem mass spectrometry. Ther. Drug Monit. 31, 683–687 (2009).
Mosmann, T. Rapid colorimetric assay for cellular growth and survival: application to proliferation and cytotoxicity assays. J. Immunol. Methods 65, 55–63 (1983).
Acknowledgements
This work was supported by the European Research Council (ERC-2008-AdG-233130-HEPCENT), INTERREG-IV-Rhin Supérieur-FEDER-Hepato-Regio-Net 2009, Agence Nationale de la Recherche (ANR-05-CEXC-008), Agence Nationale de Recherche sur le Sida 2008/354, Région Alsace, Institut National du Cancer, the Institut de Génétique et de Biologie Moléculaire et Cellulaire, Institut National de la Santé et de la Recherche Médicale, Centre National de la Recherche Scientifique, Université de Strasbourg, the US National Insititutes of Health (1K08DE020139-01A1), the UK Medical Research Council and the Wellcome Trust. We acknowledge A.-L. Morand, L. Froidevaux, A. Weiss, L. Poidevin, S. Durand and E. Soulier for excellent technical work. We thank R. Bartenschlager (University of Heidelberg) for providing Jc1 and Luc-Jc1 expression vectors, J. Ball (University of Nottingham) for UKN2A.2.4, UKN3A1.28 and UKN4.21.16 expression vectors, T. Wakita (National Institute of Infectious Diseases, Japan) for JFH1 constructs, C.M. Rice (The Rockefeller University) for Huh7.5 cells, F.V. Chisari (The Scripps Research Institute) for Huh7.5.1 cells, E. Harlow (Harvard University) for CDC2 expression plasmids and M. Tanaka (Hamamatsu University) for EphA2 expression plasmids.
Author information
Authors and Affiliations
Contributions
J.L., M.B.Z. and T.F.B. wrote the manuscript. J.L., M.B.Z., F.X., D.L., F.-L.C., J.A.M., and T.F.B. designed experiments and analyzed data. J.L., M.B.Z., F.X., C.T., I.F., L.Z., C.D., C.J.M., M.T., S.G., C.R., M.N.Z., D.L. and J.F. performed experiments. S.M.R., T.P., A.H.P., P.P. and M.D. contributed essential reagents. W.R. and O.P. performed bioinformatic analyses. J.L., B.F. and L.B. implemented and coordinated the siRNA screen. T.F.B. designed and supervised the project.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Text and Figures
Supplementary Results, Supplementary Methods and Supplementary Figures 1–12 (PDF 2570 kb)
Supplementary Table 1
Supplementary Table 1 (XLS 203 kb)
Supplementary Table 2
Supplementary Table 2 (XLS 45 kb)
Rights and permissions
About this article
Cite this article
Lupberger, J., Zeisel, M., Xiao, F. et al. EGFR and EphA2 are host factors for hepatitis C virus entry and possible targets for antiviral therapy. Nat Med 17, 589–595 (2011). https://doi.org/10.1038/nm.2341
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nm.2341
This article is cited by
-
Interferon-induced transmembrane protein-1 competitively blocks Ephrin receptor A2-mediated Epstein–Barr virus entry into epithelial cells
Nature Microbiology (2024)
-
EGFR core fucosylation, induced by hepatitis C virus, promotes TRIM40-mediated-RIG-I ubiquitination and suppresses interferon-I antiviral defenses
Nature Communications (2024)
-
Viruses exploit growth factor mechanisms to achieve augmented pathogenicity and promote tumorigenesis
Archives of Microbiology (2024)
-
Antiviral mechanisms of sorafenib against foot-and-mouth disease virus via c-RAF and AKT/PI3K pathways
Veterinary Research Communications (2024)
-
Host factor TNK2 is required for influenza virus infection
Genes & Genomics (2023)