Abstract
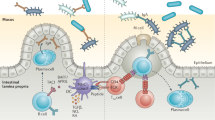
Using a systems biology approach, we discovered and dissected a three-way interaction between the immune system, the intestinal epithelium and the microbiota. We found that, in the absence of B cells, or of IgA, and in the presence of the microbiota, the intestinal epithelium launches its own protective mechanisms, upregulating interferon-inducible immune response pathways and simultaneously repressing Gata4-related metabolic functions. This shift in intestinal function leads to lipid malabsorption and decreased deposition of body fat. Network analysis revealed the presence of two interconnected epithelial-cell gene networks, one governing lipid metabolism and another regulating immunity, that were inversely expressed. Gene expression patterns in gut biopsies from individuals with common variable immunodeficiency or with HIV infection and intestinal malabsorption were very similar to those of the B cell–deficient mice, providing a possible explanation for a longstanding enigmatic association between immunodeficiency and defective lipid absorption in humans.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
References
Cerf-Bensussan, N. & Gaboriau-Routhiau, V. The immune system and the gut microbiota: friends or foes? Nat. Rev. Immunol. 10, 735–744 (2010).
Hooper, L.V. & Macpherson, A.J. Immune adaptations that maintain homeostasis with the intestinal microbiota. Nat. Rev. Immunol. 10, 159–169 (2010).
Umesaki, Y., Setoyama, H., Matsumoto, S., Imaoka, A. & Itoh, K. Differential roles of segmented filamentous bacteria and clostridia in development of the intestinal immune system. Infect. Immun. 67, 3504–3511 (1999).
Ivanov, I.I. et al. Induction of intestinal TH17 cells by segmented filamentous bacteria. Cell 139, 485–498 (2009).
Gaboriau-Routhiau, V. et al. The key role of segmented filamentous bacteria in the coordinated maturation of gut helper T cell responses. Immunity 31, 677–689 (2009).
Klaasen, H.L. et al. Apathogenic, intestinal, segmented, filamentous bacteria stimulate the mucosal immune system of mice. Infect. Immun. 61, 303–306 (1993).
Round, J.L. & Mazmanian, S.K. Inducible Foxp3+ regulatory T-cell development by a commensal bacterium of the intestinal microbiota. Proc. Natl. Acad. Sci. USA 107, 12204–12209 (2010).
Vijay-Kumar, M. et al. Metabolic syndrome and altered gut microbiota in mice lacking Toll-like receptor 5. Science 328, 228–231 (2010).
Fagarasan, S. et al. Critical roles of activation-induced cytidine deaminase in the homeostasis of gut flora. Science 298, 1424–1427 (2002).
Suzuki, K. et al. Aberrant expansion of segmented filamentous bacteria in IgA-deficient gut. Proc. Natl. Acad. Sci. USA 101, 1981–1986 (2004).
Umesaki, Y., Okada, Y., Matsumoto, S., Imaoka, A. & Setoyama, H. Segmented filamentous bacteria are indigenous intestinal bacteria that activate intraepithelial lymphocytes and induce MHC class II molecules and fucosyl asialo GM1 glycolipids on the small intestinal epithelial cells in the ex-germ-free mouse. Microbiol. Immunol. 39, 555–562 (1995).
Bry, L., Falk, P.G., Midtvedt, T. & Gordon, J.I. A model of host-microbial interactions in an open mammalian ecosystem. Science 273, 1380–1383 (1996).
Turnbaugh, P.J. et al. An obesity-associated gut microbiome with increased capacity for energy harvest. Nature 444, 1027–1031 (2006).
Gordon, J.I., Hooper, L.V., McNevin, M.S., Wong, M. & Bry, L. Epithelial cell growth and differentiation. III. Promoting diversity in the intestine: conversations between the microflora, epithelium, and diffuse GALT. Am. J. Physiol. 273, G565–G570 (1997).
Treiner, E. et al. Selection of evolutionarily conserved mucosal-associated invariant T cells by MR1. Nature 422, 164–169 (2003).
Matzinger, P. Tolerance, danger and the extended family. Annu. Rev. Immunol. 12, 991–1045 (1994).
Matzinger, P. & Kamala, T. Tissue-based class control: the other side of tolerance. Nat. Rev. Immunol. 11, 221–230 (2011).
Mizoguchi, A., Mizoguchi, E., Takedatsu, H., Blumberg, R.S. & Bhan, A.K. Chronic intestinal inflammatory condition generates IL-10–producing regulatory B cell subset characterized by CD1d upregulation. Immunity 16, 219–230 (2002).
Golovkina, T.V., Shlomchik, M., Hannum, L. & Chervonsky, A. Organogenic role of B lymphocytes in mucosal immunity. Science 286, 1965–1968 (1999).
Carragher, D.M., Kaminski, D.A., Moquin, A., Hartson, L. & Randall, T.D. A novel role for non-neutralizing antibodies against nucleoprotein in facilitating resistance to influenza virus. J. Immunol. 181, 4168–4176 (2008).
Macpherson, A.J. et al. A primitive T cell–independent mechanism of intestinal mucosal IgA responses to commensal bacteria. Science 288, 2222–2226 (2000).
Bosse, T. et al. Gata4 is essential for the maintenance of jejunal-ileal identities in the adult mouse small intestine. Mol. Cell. Biol. 26, 9060–9070 (2006).
Battle, M.A. et al. GATA4 is essential for jejunal function in mice. Gastroenterology 135, 1676–1686 (2008).
Bünger, M. et al. Genome-wide analysis of PPARα activation in murine small intestine. Physiol. Genomics 30, 192–204 (2007).
Simmen, F.A. et al. Dysregulation of intestinal crypt cell proliferation and villus cell migration in mice lacking Kruppel-like factor 9. Am. J. Physiol. Gastrointest. Liver Physiol. 292, G1757–G1769 (2007).
Falcon, A. et al. FATP2 is a hepatic fatty acid transporter and peroxisomal very long-chain acyl-CoA synthetase. Am. J. Physiol. Endocrinol. Metab. 299, E384–E393 (2010).
Lehto, M. et al. The OSBP-related protein family in humans. J. Lipid Res. 42, 1203–1213 (2001).
Hwang, B., Jeoung, N.H. & Harris, R.A. Pyruvate dehydrogenase kinase isoenzyme 4 (PDHK4) deficiency attenuates the long-term negative effects of a high-saturated fat diet. Biochem. J. 423, 243–252 (2009).
Watschinger, K. et al. Identification of the gene encoding alkylglycerol monooxygenase defines a third class of tetrahydrobiopterin-dependent enzymes. Proc. Natl. Acad. Sci. USA 107, 13672–13677 (2010).
Berger, K., Winzell, M.S., Mei, J. & Erlanson-Albertsson, C. Enterostatin and its target mechanisms during regulation of fat intake. Physiol. Behav. 83, 623–630 (2004).
Ahima, R.S. & Flier, J.S. Leptin. Annu. Rev. Physiol. 62, 413–437 (2000).
Skinner, J. et al. Construct and compare gene coexpression networks with DAPfinder and DAPview. BMC Bioinformatics 12, 286 (2011).
Nishiyama, Y. et al. Homeostatic regulation of intestinal villous epithelia by B lymphocytes. J. Immunol. 168, 2626–2633 (2002).
Degrandi, D. et al. Extensive characterization of IFN-induced GTPases mGBP1 to mGBP10 involved in host defense. J. Immunol. 179, 7729–7740 (2007).
Yanai, H., Savitsky, D., Tamura, T. & Taniguchi, T. Regulation of the cytosolic DNA-sensing system in innate immunity: a current view. Curr. Opin. Immunol. 21, 17–22 (2009).
Turnbaugh, P.J. & Gordon, J.I. The core gut microbiome, energy balance and obesity. J. Physiol. (Lond.) 587, 4153–4158 (2009).
Hughes, W.S., Cerda, J.J., Holtzapple, P. & Brooks, F.P. Primary hypogammaglobulinemia and malabsorption. Ann. Intern. Med. 74, 903–910 (1971).
Mannon, P.J. et al. Excess IL-12 but not IL-23 accompanies the inflammatory bowel disease associated with common variable immunodeficiency. Gastroenterology 131, 748–756 (2006).
Agarwal, S. & Mayer, L. Pathogenesis and treatment of gastrointestinal disease in antibody deficiency syndromes. J. Allergy Clin. Immunol. 124, 658–664 (2009).
Koch, J. et al. Steatorrhea: a common manifestation in patients with HIV/AIDS. Nutrition 12, 507–510 (1996).
Ullrich, R., Riecken, E.O. & Zeitz, M. Human immunodeficiency virus–induced enteropathy. Immunol. Res. 10, 456–464 (1991).
Sanford, J.P., Favour, C.B. & Tribeman, MS. Absence of serum gamma globulins in an adult. N. Engl. J. Med. 250, 1027–1029 (1954).
Owens, S.R. & Greenson, J.K. The pathology of malabsorption: current concepts. Histopathology 50, 64–82 (2007).
Malamut, G. et al. The enteropathy associated with common variable immunodeficiency: the delineated frontiers with celiac disease. Am. J. Gastroenterol. 105, 2262–2275 (2010).
Lerner, P. et al. The gut mucosal viral reservoir in HIV-infected patients is not the major source of rebound plasma viremia following interruption of highly active antiretroviral therapy. J. Virol. 85, 4772–4782 (2011).
Crawford, A., Macleod, M., Schumacher, T., Corlett, L. & Gray, D. Primary T cell expansion and differentiation in vivo requires antigen presentation by B cells. J. Immunol. 176, 3498–3506 (2006).
Wojciechowski, W. et al. Cytokine-producing effector B cells regulate type 2 immunity to H. polygyrus. Immunity 30, 421–433 (2009).
Cong, Y., Feng, T., Fujihashi, K., Schoeb, T.R. & Elson, C.O. A dominant, coordinated T regulatory cell–IgA response to the intestinal microbiota. Proc. Natl. Acad. Sci. USA 106, 19256–19261 (2009).
Peterson, D.A., McNulty, N.P., Guruge, J.L. & Gordon, J.I. IgA response to symbiotic bacteria as a mediator of gut homeostasis. Cell Host Microbe 2, 328–339 (2007).
Obata, T. et al. Indigenous opportunistic bacteria inhabit mammalian gut-associated lymphoid tissues and share a mucosal antibody-mediated symbiosis. Proc. Natl. Acad. Sci. USA 107, 7419–7424 (2010).
Scamurra, R.W. et al. Mucosal plasma cell repertoire during HIV-1 infection. J. Immunol. 169, 4008–4016 (2002).
Kotler, D.P., Scholes, J.V. & Tierney, A.R. Intestinal plasma cell alterations in acquired immunodeficiency syndrome. Dig. Dis. Sci. 32, 129–138 (1987).
Shulzhenko, N., Morgun, A. & Matzinger, P. Spontaneous mutation in the Cd79b gene leads to a block in B-lymphocyte development at the C (early pre-B) stage. Genes Immun. 10, 722–726 (2009).
Cline, M.S. et al. Integration of biological networks and gene expression data using Cytoscape. Nat. Protoc. 2, 2366–2382 (2007).
Bader, G.D. & Hogue, C.W. An automated method for finding molecular complexes in large protein interaction networks. BMC Bioinformatics 4, 2 (2003).
Barman, M. et al. Enteric salmonellosis disrupts the microbial ecology of the murine gastrointestinal tract. Infect. Immun. 76, 907–915 (2008).
Cole, J.R. et al. The Ribosomal Database Project (RDP-II): previewing a new autoaligner that allows regular updates and the new prokaryotic taxonomy. Nucleic Acids Res. 31, 442–443 (2003).
Schloss, P.D. et al. Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl. Environ. Microbiol. 75, 7537–7541 (2009).
Acknowledgements
This research was supported in part by the Intramural Research Program of the NIAID, NIH. We thank T. Myers, Q. Su and A. Godinez of the NIAID microarray facility for excellent technical support; R. Schwartz for providing funding for germ-free re-derivation; W. Strober for help with human subjects; C. Jones for technical support of bacterial pyrosequencing; T.D. Randall (Trudeau Institute) for AID/μS mice; S. Epstein (US Food and Drug Administration), J. Misplon (US Food and Drug Administration) and D.P. Huston (Texas A&M Health Science Center) for IgAKO mice; D. Kaiserlian (Institut National de la Santé et de la Recherche Médicale) and R. Blumberg (Harvard University) for providing the MODE-K cell line; R. Varma for help in experiments; Laboratory of Cellular and Molecular Immunology members and M. Sterman Dolnikoff, I. Shmulevich and A. Dzutsev for discussions and suggestions; Y. Kotliarov for help with graphics; S. Varma for help finding human gene homologues; J. Coursen for excellent technical support; B. Epstein for assistance with software programming; and the personnel of NIH mouse facilities in buildings 4 and 6B. The Cincinnati Mouse Metabolic Phenotyping Center is supported by NIH grant U24 DK059630 and National Gnotobiotic Rodent Resource Center at the University of North Carolina is supported by grant P40RR018603.
Author information
Authors and Affiliations
Contributions
N.S. and A.M. conceived the original idea, designed the study, conducted most of the experiments, analyzed the data and wrote the manuscript. W.H. analyzed microbiome data and drafted the related part of the manuscript. M.B. provided Gata4KOvil mice and did some Gata4-related experiments. M.Y. and L.M. collected duodenal biopsies and performed clinical evaluation of human subjects. O.G. performed and analyzed experiments related to lipid metabolism. M.O. performed histological evaluation of mouse samples. A.J.M. and K.D.M. re-derived germ-free JhKO mice and collected organs. C.F.-L. supervised microbiome evaluation and drafted the related part of the manuscript. P.M. supervised the whole study and wrote the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Text and Figures
Supplementary Data, Supplementary Methods and Supplementary Figures 1–10 (PDF 1078 kb)
Supplementary Tables 1–4
Supplementary Tables 1–4 (XLS 135 kb)
Rights and permissions
About this article
Cite this article
Shulzhenko, N., Morgun, A., Hsiao, W. et al. Crosstalk between B lymphocytes, microbiota and the intestinal epithelium governs immunity versus metabolism in the gut. Nat Med 17, 1585–1593 (2011). https://doi.org/10.1038/nm.2505
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nm.2505
This article is cited by
-
Transkingdom Network Analysis (TkNA): a systems framework for inferring causal factors underlying host–microbiota and other multi-omic interactions
Nature Protocols (2024)
-
Cardiovascular disease risk assessment through sensing the circulating microbiome with perovskite quantum dots leveraging deep learning models for bacterial species selection
Microchimica Acta (2024)
-
Gut microbiota and acute kidney injury: immunological crosstalk link
International Urology and Nephrology (2023)
-
Interferon-Driven Immune Dysregulation in Common Variable Immunodeficiency–Associated Villous Atrophy and Norovirus Infection
Journal of Clinical Immunology (2023)
-
A narrative review of the moderating effects and repercussion of exercise intervention on osteoporosis: ingenious involvement of gut microbiota and its metabolites
Journal of Translational Medicine (2022)