Abstract
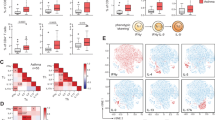
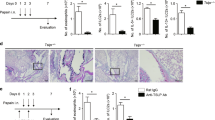
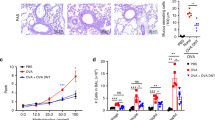
Allergic asthma is a T helper type 2 (TH2)-dominated disease of the lung. In people with asthma, a fraction of CD4+ T cells express the CX3CL1 receptor, CX3CR1, and CX3CL1 expression is increased in airway smooth muscle, lung endothelium and epithelium upon allergen challenge. Here we found that untreated CX3CR1-deficient mice or wild-type (WT) mice treated with CX3CR1-blocking reagents show reduced lung disease upon allergen sensitization and challenge. Transfer of WT CD4+ T cells into CX3CR1-deficient mice restored the cardinal features of asthma, and CX3CR1-blocking reagents prevented airway inflammation in CX3CR1-deficient recipients injected with WT TH2 cells. We found that CX3CR1 signaling promoted TH2 survival in the inflamed lungs, and injection of B cell leukemia/lymphoma-2 protein (BCl-2)-transduced CX3CR1-deficient TH2 cells into CX3CR1-deficient mice restored asthma. CX3CR1-induced survival was also observed for TH1 cells upon airway inflammation but not under homeostatic conditions or upon peripheral inflammation. Therefore, CX3CR1 and CX3CL1 may represent attractive therapeutic targets in asthma.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
References
Wills-Karp, M. Immunologic basis of antigen-induced airway hyperresponsiveness. Annu. Rev. Immunol. 17, 255–281 (1999).
Robinson, D.S. et al. Predominant TH2-like bronchoalveolar T-lymphocyte population in atopic asthma. N. Engl. J. Med. 326, 298–304 (1992).
Bazan, J.F. et al. A new class of membrane-bound chemokine with a CX3C motif. Nature 385, 640–644 (1997).
Papadopoulos, E.J. et al. Fractalkine, a CX3C chemokine, is expressed by dendritic cells and is up-regulated upon dendritic cell maturation. Eur. J. Immunol. 29, 2551–2559 (1999).
Muehlhoefer, A. et al. Fractalkine is an epithelial and endothelial cell-derived chemoattractant for intraepithelial lymphocytes in the small intestinal mucosa. J. Immunol. 164, 3368–3376 (2000).
Nanki, T. et al. Inhibition of fractalkine ameliorates murine collagen-induced arthritis. J. Immunol. 173, 7010–7016 (2004).
Yajima, N. et al. Elevated levels of soluble fractalkine in active systemic lupus erythematosus: potential involvement in neuropsychiatric manifestations. Arthritis Rheum. 52, 1670–1675 (2005).
Imai, T. et al. Identification and molecular characterization of fractalkine receptor CX3CR1, which mediates both leukocyte migration and adhesion. Cell 91, 521–530 (1997).
Geissmann, F., Jung, S. & Littman, D.R. Blood monocytes consist of two principal subsets with distinct migratory properties. Immunity 19, 71–82 (2003).
Jung, S. et al. Analysis of fractalkine receptor CX(3)CR1 function by targeted deletion and green fluorescent protein reporter gene insertion. Mol. Cell. Biol. 20, 4106–4114 (2000).
Foussat, A. et al. Fractalkine receptor expression by T lymphocyte subpopulations and in vivo production of fractalkine in human. Eur. J. Immunol. 30, 87–97 (2000).
Fraticelli, P. et al. Fractalkine (CX3CL1) as an amplification circuit of polarized TH1 responses. J. Clin. Invest. 107, 1173–1181 (2001).
Nishimura, M. et al. Dual functions of fractalkine/CX3C ligand 1 in trafficking of perforin+/granzyme B+ cytotoxic effector lymphocytes that are defined by CX3CR1 expression. J. Immunol. 168, 6173–6180 (2002).
Lesnik, P., Haskell, C.A. & Charo, I.F. Decreased atherosclerosis in Cx3cr1−/− mice reveals a role for fractalkine in atherogenesis. J. Clin. Invest. 111, 333–340 (2003).
Inoue, A. et al. Antagonist of fractalkine (CX3CL1) delays the initiation and ameliorates the progression of lupus nephritis in MRL/lpr mice. Arthritis Rheum. 52, 1522–1533 (2005).
Suzuki, F. et al. Inhibition of CX3CL1 (fractalkine) improves experimental autoimmune myositis in SJL/J mice. J. Immunol. 175, 6987–6996 (2005).
Echigo, T., Hasegawa, M., Shimada, Y., Takehara, K. & Sato, S. Expression of fractalkine and its receptor, CX3CR1, in atopic dermatitis: possible contribution to skin inflammation. J. Allergy Clin. Immunol. 113, 940–948 (2004).
Rimaniol, A.C. et al. The CX3C chemokine fractalkine in allergic asthma and rhinitis. J. Allergy Clin. Immunol. 112, 1139–1146 (2003).
El-Shazly, A. et al. Fraktalkine produced by airway smooth muscle cells contributes to mast cell recruitment in asthma. J. Immunol. 176, 1860–1868 (2006).
Tremblay, K. et al. Association study between the CX3CR1 gene and asthma. Genes Immun. 7, 632–639 (2006).
Depner, M. et al. CX3CR1 Polymorphisms are associated with atopy but not asthma in German children. Int. Arch. Allergy Immunol. 144, 91–94 (2007).
Cohn, L., Homer, R.J., Marinov, A., Rankin, J. & Bottomly, K. Induction of airway mucus production By T helper 2 (TH2) cells: a critical role for interleukin 4 in cell recruitment but not mucus production. J. Exp. Med. 186, 1737–1747 (1997).
Chvatchko, Y. et al. A key role for CC chemokine receptor 4 in lipopolysaccharide-induced endotoxic shock. J. Exp. Med. 191, 1755–1764 (2000).
Humbles, A.A. et al. The murine CCR3 receptor regulates both the role of eosinophils and mast cells in allergen-induced airway inflammation and hyperresponsiveness. Proc. Natl. Acad. Sci. USA 99, 1479–1484 (2002).
Conroy, D.M. et al. CCR4 blockade does not inhibit allergic airways inflammation. J. Leukoc. Biol. 74, 558–563 (2003).
Kawasaki, S. et al. Intervention of thymus and activation-regulated chemokine attenuates the development of allergic airway inflammation and hyperresponsiveness in mice. J. Immunol. 166, 2055–2062 (2001).
Mikhak, Z. et al. Contribution of CCR4 and CCR8 to antigen-specific TH2 cell trafficking in allergic pulmonary inflammation. J. Allergy Clin. Immunol. 123, 67–73.e3 (2009).
Chung, C.D. et al. CCR8 is not essential for the development of inflammation in a mouse model of allergic airway disease. J. Immunol. 170, 581–587 (2003).
Chensue, S.W. et al. Aberrant in vivo T helper type 2 cell response and impaired eosinophil recruitment in CC chemokine receptor 8 knockout mice. J. Exp. Med. 193, 573–584 (2001).
Goya, I. et al. Absence of CCR8 does not impair the response to ovalbumin-induced allergic airway disease. J. Immunol. 170, 2138–2146 (2003).
Gonzalo, J.A. et al. Critical involvement of the chemotactic axis CXCR4/stromal cell-derived factor-1α in the inflammatory component of allergic airway disease. J. Immunol. 165, 499–508 (2000).
Lukacs, N.W., Berlin, A., Schols, D., Skerlj, R.T. & Bridger, G.J. AMD3100, a CxCR4 antagonist, attenuates allergic lung inflammation and airway hyperreactivity. Am. J. Pathol. 160, 1353–1360 (2002).
Liu, X. et al. The CC chemokine ligand 2 (CCL2) mediates fibroblast survival through IL-6. Am. J. Respir. Cell Mol. Biol. 37, 121–128 (2007).
Hippe, A., Homey, B. & Mueller-Homey, A. Chemokines. Recent Results Cancer Res. 180, 35–50 (2010).
Nie, Y., Han, Y.C. & Zou, Y.R. CXCR4 is required for the quiescence of primitive hematopoietic cells. J. Exp. Med. 205, 777–783 (2008).
Linge, H.M. et al. The human CXC chemokine granulocyte chemotactic protein 2 (GCP-2)/CXCL6 possesses membrane-disrupting properties and is antibacterial. Antimicrob. Agents Chemother. 52, 2599–2607 (2008).
Linge, H.M. et al. The antibacterial chemokine MIG/CXCL9 is constitutively expressed in epithelial cells of the male urogenital tract and is present in seminal plasma. J. Interferon Cytokine Res. 28, 191–196 (2008).
Yang, D. et al. Many chemokines including CCL20/MIP-3alpha display antimicrobial activity. J. Leukoc. Biol. 74, 448–455 (2003).
Benamar, K., Geller, E.B. & Adler, M.W. Elevated level of the proinflammatory chemokine, RANTES/CCL5, in the periaqueductal grey causes hyperalgesia in rats. Eur. J. Pharmacol. 592, 93–95 (2008).
Milligan, E.D., Sloane, E.M. & Watkins, L.R. Glia in pathological pain: a role for fractalkine. J. Neuroimmunol. 198, 113–120 (2008).
Miller, R.J., Jung, H., Bhangoo, S.K. & White, F.A. Cytokine and chemokine regulation of sensory neuron function. Handb. Exp. Pharmacol. 194, 417–449 (2009).
Meucci, O. et al. Chemokines regulate hippocampal neuronal signaling and gp120 neurotoxicity. Proc. Natl. Acad. Sci. USA 95, 14500–14505 (1998).
Landsman, L. et al. CX3CR1 is required for monocyte homeostasis and atherogenesis by promoting cell survival. Blood 113, 963–972 (2008).
Garin, A., Pellet, P., Deterre, P., Debre, P. & Combadiere, C. Cloning and functional characterization of the human fractalkine receptor promoter regions. Biochem. J. 368, 753–760 (2002).
Chen, S., Luo, D., Streit, W.J. & Harrison, J.K. TGF-β1 upregulates CX3CR1 expression and inhibits fractalkine-stimulated signaling in rat microglia. J. Neuroimmunol. 133, 46–55 (2002).
Ramos, M.V. et al. Interleukin-10 and interferon-γ modulate surface expression of fractalkine-receptor (CX(3)CR1) via PI3K in monocytes. Immunology 129, 600–609 (2010).
El Biaze, M. et al. T cell activation, from atopy to asthma: more a paradox than a paradigm. Allergy 58, 844–853 (2003).
Boniface, S. et al. Assessment of T lymphocyte cytokine production in induced sputum from asthmatics: a flow cytometry study. Clin. Exp. Allergy 33, 1238–1243 (2003).
Cho, S.H., Stanciu, L.A., Holgate, S.T. & Johnston, S.L. Increased interleukin-4, interleukin-5, and interferon-γ in airway CD4+ and CD8+ T cells in atopic asthma. Am. J. Respir. Crit. Care Med. 171, 224–230 (2005).
Mamessier, E. et al. T-cell activation during exacerbations: a longitudinal study in refractory asthma. Allergy 63, 1202–1210 (2008).
Lopez Vina, A. Severe asthma refractory to treatment: concepts and realities. Arch. Bronconeumol. 42, 20–25 (2006).
O'Byrne, P.M. The demise of anti IL-5 for asthma, or not. Am. J. Respir. Crit. Care. Med. 176, 1059–1060 (2007).
Wang, Q. et al. CD4 promotes breadth in the TCR repertoire. J. Immunol. 167, 4311–4320 (2001).
Kheradmand, F. et al. A protease-activated pathway underlying TH cell type 2 activation and allergic lung disease. J. Immunol. 169, 5904–5911 (2002).
Mougneau, E. et al. Expression cloning of a Leishmania major protective T cell antigen. Science 268, 563–566 (1995).
Hamelmann, E. et al. Primary prevention of allergy: avoiding risk or providing protection? Clin. Exp. Allergy 38, 233–245 (2008).
Verhasselt, V. et al. Breast milk-mediated transfer of an antigen induces tolerance and protection from allergic asthma. Nat. Med. 14, 170–175 (2008).
Honda, K., Marquillies, P., Capron, M. & Dombrowicz, D. Peroxisome proliferator-activated receptor gamma is expressed in airways and inhibits features of airway remodeling in a mouse asthma model. J. Allergy Clin. Immunol. 113, 882–888 (2004).
Humbles, A.A. et al. A critical role for eosinophils in allergic airways remodeling. Science 305, 1776–1779 (2004).
Acknowledgements
We thank N. Guy, V. Thieffin and A. Barbot for animal care, J. Cazareth and F. Larbret for cell sorting and S. Kirschnek (Institute for Medical Microbiology, Immunology and Hygiene, Technical University Munich) for providing the MIG-mBcl-2 and MIGR1 vectors. IFN-γ–specific antibody R4-6A2 and IL-4–specific antibody 11B11 were kind gifts from DNAX Research Institute. Cx3cr1GFP/GFP mice were a gift from S. Jung (Weizmann Institute of Science, Immunology Department). This research was supported by the Fondation pour la Recherche Medical, the Agence Nationale de la Recherche, the French Society of Allergology, the French Pneumologie Society, the Genavie foundation and the Pays de la Loire and Provence Alpes Cote d'Azur regional councils.
Author information
Authors and Affiliations
Contributions
C.M. and V.B. conducted most of the experiments and contributed to data analysis. A.K., S.F. and D.D. measured lung resistance and compliance and performed lung histology. V.M. provided technical help. D.L., M.L., Y.L. and A.M. performed experiments with HDM. E.H. and R.C. contributed to early experiments aimed at monitoring chemokine receptor expression in lung T cells. N.G. and V.J. wrote the manuscript. V.J. conceived of and directed the project.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–14 (PDF 2064 kb)
Rights and permissions
About this article
Cite this article
Mionnet, C., Buatois, V., Kanda, A. et al. CX3CR1 is required for airway inflammation by promoting T helper cell survival and maintenance in inflamed lung. Nat Med 16, 1305–1312 (2010). https://doi.org/10.1038/nm.2253
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nm.2253
This article is cited by
-
CX3CR1 modulates SLE-associated glomerulonephritis and cardiovascular disease in MRL/lpr mice
Inflammation Research (2023)
-
Immune phenotypes that are associated with subsequent COVID-19 severity inferred from post-recovery samples
Nature Communications (2022)
-
RSV pre-fusion F protein enhances the G protein antibody and anti-infectious responses
npj Vaccines (2022)
-
Regulation and function of CX3CR1 and its ligand CX3CL1 in kidney disease
Cell and Tissue Research (2021)
-
CX3CR1 positively regulates BCR signaling coupled with cell metabolism via negatively controlling actin remodeling
Cellular and Molecular Life Sciences (2020)