Abstract
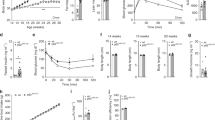
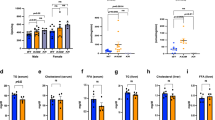
Obesity and metabolic syndrome are increasingly recognized as major risk factors for cardiovascular disease. Herein we show that Krüppel-like transcription factor 5 (KLF5) is a crucial regulator of energy metabolism. Klf5+/− mice were resistant to high fat–induced obesity, hypercholesterolemia and glucose intolerance, despite consuming more food than wild-type mice. This may in part reflect their enhanced energy expenditure. Expression of the genes involved in lipid oxidation and energy uncoupling, including those encoding carnitine-palmitoyl transferase-1b (Cpt1b) and uncoupling proteins 2 and 3 (Ucp2 and Ucp3), was upregulated in the soleus muscles of Klf5+/− mice. Under basal conditions, KLF5 modified with small ubiquitin-related modifier (SUMO) proteins was associated with transcriptionally repressive regulatory complexes containing unliganded peroxisome proliferator–activated receptor-δ (PPAR-δ) and co-repressors and thus inhibited Cpt1b, Ucp2 and Ucp3 expression. Upon agonist stimulation of PPAR-δ, KLF5 was deSUMOylated, and became associated with transcriptional activation complexes containing both the liganded PPAR-δ and CREB binding protein (CBP). This activation complex increased the expression of Cpt1b, Ucp2 and Ucp3. Thus, SUMOylation seems to be a molecular switch affecting function of KLF5 and the transcriptional regulatory programs governing lipid metabolism.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
References
Lowell, B.B. & Spiegelman, B.M. Towards a molecular understanding of adaptive thermogenesis. Nature 404, 652–660 (2000).
Weigle, D.S. Appetite and the regulation of body composition. FASEB J. 8, 302–310 (1994).
Smith, A.G. & Muscat, G.E. Skeletal muscle and nuclear hormone receptors: implications for cardiovascular and metabolic disease. Int. J. Biochem. Cell Biol. 37, 2047–2063 (2005).
Ukropcova, B. et al. Dynamic changes in fat oxidation in human primary myocytes mirror metabolic characteristics of the donor. J. Clin. Invest. 115, 1934–1941 (2005).
Kelley, D.E. Skeletal muscle fat oxidation: timing and flexibility are everything. J. Clin. Invest. 115, 1699–1702 (2005).
Spiegelman, B.M. & Heinrich, R. Biological control through regulated transcriptional coactivators. Cell 119, 157–167 (2004).
Lin, J., Handschin, C. & Spiegelman, B.M. Metabolic control through the PGC-1 family of transcription coactivators. Cell Metab. 1, 361–370 (2005).
Rawson, R.B. The SREBP pathway. Nat. Rev. Mol. Cell Biol. 4, 631–640 (2003).
Evans, R.M., Barish, G.D. & Wang, Y.X. PPARs and the complex journey to obesity. Nat. Med. 10, 355–361 (2004).
Mootha, V.K. et al. Errα and Gabpa/b specify PGC-1α–dependent oxidative phosphorylation gene expression that is altered in diabetic muscle. Proc. Natl. Acad. Sci. USA 101, 6570–6575 (2004).
Wolfrum, C., Asilmaz, E., Luca, E., Friedman, J.M. & Stoffel, M. Foxa2 regulates lipid metabolism and ketogenesis in the liver during fasting and in diabetes. Nature 432, 1027–1032 (2004).
Schuler, M. et al. PGC1α expression is controlled in skeletal muscles by PPARβ, whose ablation results in fiber-type switching, obesity, and type 2 diabetes. Cell Metab. 4, 407–414 (2006).
Dressel, U. et al. The peroxisome proliferator–activated receptor β/δ agonist, GW501516, regulates the expression of genes involved in lipid catabolism and energy uncoupling in skeletal muscle cells. Mol. Endocrinol. 17, 2477–2493 (2003).
Tanaka, T. et al. Activation of peroxisome proliferator–activated receptor δ induces fatty acid β-oxidation in skeletal muscle and attenuates metabolic syndrome. Proc. Natl. Acad. Sci. USA 100, 15924–15929 (2003).
Wang, Y.X. et al. Peroxisome proliferator–activated receptor δ activates fat metabolism to prevent obesity. Cell 113, 159–170 (2003).
Wang, Y.X. et al. Regulation of muscle fiber type and running endurance by PPARδ. PLoS Biol. 2, e294 (2004).
Lee, C.H. et al. Peroxisome proliferator–activated receptor δ promotes very low-density lipoprotein–derived fatty acid catabolism in the macrophage. Proc. Natl. Acad. Sci. USA 103, 2434–2439 (2006).
Krogsdam, A.M. et al. Nuclear receptor corepressor–dependent repression of peroxisome proliferator–activated receptor δ–mediated transactivation. Biochem. J. 363, 157–165 (2002).
Shi, Y., Hon, M. & Evans, R.M. The peroxisome proliferator–activated receptor δ, an integrator of transcriptional repression and nuclear receptor signaling. Proc. Natl. Acad. Sci. USA 99, 2613–2618 (2002).
Hay, R.T. SUMO: a history of modification. Mol. Cell 18, 1–12 (2005).
Gill, G. Something about SUMO inhibits transcription. Curr. Opin. Genet. Dev. 15, 536–541 (2005).
Pascual, G. et al. A SUMOylation-dependent pathway mediates transrepression of inflammatory response genes by PPAR-γ. Nature 437, 759–763 (2005).
Eaton, E.M. & Sealy, L. Modification of CCAAT/enhancer-binding protein-β by the small ubiquitin-like modifier (SUMO) family members, SUMO-2 and SUMO-3. J. Biol. Chem. 278, 33416–33421 (2003).
Hirano, Y., Murata, S., Tanaka, K., Shimizu, M. & Sato, R. Sterol regulatory element-binding proteins are negatively regulated through SUMO-1 modification independent of the ubiquitin/26 S proteasome pathway. J. Biol. Chem. 278, 16809–16819 (2003).
Haldar, S.M., Ibrahim, O.A. & Jain, M.K. Kruppel-like factors (KLFs) in muscle biology. J. Mol. Cell. Cardiol. 43, 1–10 (2007).
Fujiu, K. et al. Synthetic retinoid Am80 suppresses smooth muscle phenotypic modulation and in-stent neointima formation by inhibiting KLF5. Circ. Res. 97, 1132–1141 (2005).
Shindo, T. et al. Kruppel-like zinc-finger transcription factor KLF5/BTEB2 is a target for angiotensin II signaling and an essential regulator of cardiovascular remodeling. Nat. Med. 8, 856–863 (2002).
Oishi, Y. et al. Kruppel-like transcription factor KLF5 is a key regulator of adipocyte differentiation. Cell Metab. 1, 27–39 (2005).
Shi, H., Zhang, Z., Wang, X., Liu, S. & Teng, C.T. Isolation and characterization of a gene encoding human Kruppel-like factor 5 (IKLF): binding to the CAAT/GT box of the mouse lactoferrin gene promoter. Nucleic Acids Res. 27, 4807–4815 (1999).
Watanabe, N. et al. BTEB2, a Kruppel-like transcription factor, regulates expression of the SMemb/Nonmuscle myosin heavy chain B (SMemb/NMHC-B) gene. Circ. Res. 85, 182–191 (1999).
Brandt, J.M., Djouadi, F. & Kelly, D.P. Fatty acids activate transcription of the muscle carnitine palmitoyltransferase I gene in cardiac myocytes via the peroxisome proliferator–activated receptor α. J. Biol. Chem. 273, 23786–23792 (1998).
Medvedev, A.V., Snedden, S.K., Raimbault, S., Ricquier, D. & Collins, S. Transcriptional regulation of the mouse uncoupling protein-2 gene. Double E-box motif is required for peroxisome proliferator–activated receptor-γ–dependent activation. J. Biol. Chem. 276, 10817–10823 (2001).
Pecqueur, C. et al. Uncoupling protein 2, in vivo distribution, induction upon oxidative stress and evidence for translational regulation. J. Biol. Chem. 276, 8705–8712 (2001).
van der Leij, F.R. et al. Structural and functional genomics of the CPT1B gene for muscle-type carnitine palmitoyltransferase I in mammals. J. Biol. Chem. 277, 26994–27005 (2002).
Dang, D.T., Zhao, W., Mahatan, C.S., Geiman, D.E. & Yang, V.W. Opposing effects of Kruppel-like factor 4 (gut-enriched Kruppel-like factor) and Kruppel-like factor 5 (intestinal-enriched Kruppel-like factor) on the promoter of the Kruppel-like factor 4 gene. Nucleic Acids Res. 30, 2736–2741 (2002).
Gong, L. & Yeh, E.T. Characterization of a family of nucleolar SUMO-specific proteases with preference for SUMO-2 or SUMO-3. J. Biol. Chem. 281, 15869–15877 (2006).
Tatham, M.H. et al. Polymeric chains of SUMO-2 and SUMO-3 are conjugated to protein substrates by SAE1/SAE2 and Ubc9. J. Biol. Chem. 276, 35368–35374 (2001).
Perdomo, J., Verger, A., Turner, J. & Crossley, M. Role for SUMO modification in facilitating transcriptional repression by BKLF. Mol. Cell. Biol. 25, 1549–1559 (2005).
Chou, C.C. et al. Small ubiquitin-like modifier modification regulates the DNA binding activity of glial cell missing Drosophila homolog a. J. Biol. Chem. 282, 27239–27249 (2007).
Kerner, J. & Hoppel, C. Fatty acid import into mitochondria. Biochim. Biophys. Acta 1486, 1–17 (2000).
Sebastian, D., Herrero, L., Serra, D., Asins, G. & Hegardt, F.G. CPT I overexpression protects L6E9 muscle cells from fatty acid–induced insulin resistance. Am. J. Physiol. Endocrinol. Metab. 292, E677–E686 (2007).
Dobbins, R.L. et al. Prolonged inhibition of muscle carnitine palmitoyltransferase-1 promotes intramyocellular lipid accumulation and insulin resistance in rats. Diabetes 50, 123–130 (2001).
Vidal-Puig, A.J. et al. Energy metabolism in uncoupling protein 3 gene knockout mice. J. Biol. Chem. 275, 16258–16266 (2000).
Bezaire, V., Seifert, E.L. & Harper, M.E. Uncoupling protein-3: clues in an ongoing mitochondrial mystery. FASEB J. 21, 312–324 (2007).
Brand, M.D. & Esteves, T.C. Physiological functions of the mitochondrial uncoupling proteins UCP2 and UCP3. Cell Metab. 2, 85–93 (2005).
Rosenfeld, M.G., Lunyak, V.V. & Glass, C.K. Sensors and signals: a coactivator/corepressor/epigenetic code for integrating signal-dependent programs of transcriptional response. Genes Dev. 20, 1405–1428 (2006).
Barish, G.D., Narkar, V.A. & Evans, R.M. PPARδ: a dagger in the heart of the metabolic syndrome. J. Clin. Invest. 116, 590–597 (2006).
Lee, C.H. et al. PPARδ regulates glucose metabolism and insulin sensitivity. Proc. Natl. Acad. Sci. USA 103, 3444–3449 (2006).
Acknowledgements
We gratefully acknowledge N. Yamanaka, M. Hayashi, Y. Tani and E. Magoshi for their excellent technical assistance. We thank M. Yamada (Gunma University) for providing pGL3-UCP2, C. Glass (University of California, San Diego) for providing FLAG-tagged cDNA encoding mouse NCoR, R. Evans (Salk Institute for Biological Studies) for providing cDNA encoding human SMRT and the Gal4-SMRT expression construct, R. Hay (University of Dundee) for providing pcDNA3-HA-SUMO1 and pcDNA3-HA-SUMO2, E. Yeh (University of Texas–Houston Health Science) for providing pcDNA3-SENP1, and P. Grimaldi (Universite de Nice-Sophia Antipolis) for providing pcDNA3-PPARδ. This study was supported in part by grants-in-aid from the Ministry of Education, Culture, Sports, Science and Technology, Japan (to R.N., I.M. and Y.O.), a research grant from the National Institute of Biomedical Innovation, Japan and Special Coordination Funds for Promoting Science and Technology from Japan Science and Technology Agency (to R.N.), and research grants from Japan Science and Technology Agency, NOVARTIS Foundation for the Promotion of Science, Kato Memorial Bioscience Foundation, Takeda Science Foundation, Cell Science Research Foundation and Tokyo Biochemical Research Foundation (to I.M.).
Author information
Authors and Affiliations
Contributions
Y.O. developed the project, performed most of the experiments and data analysis and wrote the manuscript. I.M. developed and supervised the project, designed experiments, performed data analysis and wrote the manuscript. M.O., T. Kubota. and N.K. contributed to the mouse physiology studies. K.F. and K.M. contributed to the in vitro studies. K.T. and T. Kadowaki supervised the project and reviewed the manuscript. R.N. directed the project and reviewed the manuscript.
Corresponding authors
Supplementary information
Supplementary Text and Figures
Supplementary Figs. 1–7 and Supplementary Methods (PDF 3311 kb)
Rights and permissions
About this article
Cite this article
Oishi, Y., Manabe, I., Tobe, K. et al. SUMOylation of Krüppel-like transcription factor 5 acts as a molecular switch in transcriptional programs of lipid metabolism involving PPAR-δ. Nat Med 14, 656–666 (2008). https://doi.org/10.1038/nm1756
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nm1756
This article is cited by
-
The KLF7/PFKL/ACADL axis modulates cardiac metabolic remodelling during cardiac hypertrophy in male mice
Nature Communications (2023)
-
Targeting protein modifications in metabolic diseases: molecular mechanisms and targeted therapies
Signal Transduction and Targeted Therapy (2023)
-
SENP2 regulates mitochondrial function and insulin secretion in pancreatic β cells
Experimental & Molecular Medicine (2022)
-
PPAR control of metabolism and cardiovascular functions
Nature Reviews Cardiology (2021)
-
Eosinophil function in adipose tissue is regulated by Krüppel-like factor 3 (KLF3)
Nature Communications (2020)