Abstract
It is assumed that a timely mass administration of antiviral drugs, backed by quarantines and social distancing, could contain a nascent influenza epidemic at its source, provided that the first clusters of cases were localized within a short time. However, effective routine surveillance may be impossible in countries lacking basic public health resources. For a global containment strategy to be successful, low-cost, easy-to-use handheld units that permit decentralized testing would be vital. Here we present a microfluidic platform that can detect the highly pathogenic avian influenza virus H5N1 in a throat swab sample by using magnetic forces to manipulate a free droplet containing superparamagnetic particles. In a sequential process, the viral RNA is isolated, purified, preconcentrated by 50,000% and subjected to ultrafast real-time RT-PCR. Compared to commercially available tests, the bioassay is equally sensitive and is 440% faster and 2,000–5,000% cheaper.
Similar content being viewed by others
Main
Highly pathogenic avian influenza (HPAI) H5N1 is now entrenched in Southeast Asia, with sporadic human infections resulting from either direct contacts with infected birds1 or limited human-to-human transmissions2. Should the virus improve its ability to achieve efficient and sustained transmission between humans by antigenic drift and/or shift3, a pandemic could emerge4.
Based on stochastic models that emphasize the idea of an early intervention, the WHO containment plan5 aims to stop an epidemic locally in order to prevent a global disaster. In the best-case scenarios, an incipient epidemic is contained if a new virus is spotted either within the first 20 cases6 or within up to 21 d after the first case emerges7. Hence the success of the plan depends solely on how fast healthcare professionals can identify and treat infected persons.
The regular monitoring of a potential outbreak may be a Gordian knot in some of the developing Asian8,9,10, Middle Eastern11 and African countries, where adequate instrumentation and/or diagnostic test kits for sample collection, isolation, RT-PCR12 and gel electrophoresis are either unaffordable or restricted to a few laboratories. Clearly, to halt a pandemic emerging in low-resource settings, there is a need for low-cost, easy-to-use handheld units that decentralize testing. However, no microfluidic diagnostic devices are currently available to tackle (re)emerging infectious or parasitic diseases such as influenza, HIV/AIDS, cholera, malaria, tuberculosis or measles13.
An ideal microfluidic concept integrates a set of generic components, such as an interface for introducing preferably liquid samples and reagents, a technique for isolation, purification and preconcentration of samples and/or reaction products, a method for moving, merging and mixing these liquids, and a miniaturized detection system14. Although impressive solutions have been demonstrated for most of the single components, their overall integration, is challenging, and most of the previous preparations lack the ability to process complex real-world samples15.
With a few exceptions16, the sample preparation is performed off-chip. Without purification and/or preconcentration, biological samples such as whole blood, urine, saliva or feces cannot be analyzed in a microfluidic environment. Solids, particulates, air bubbles, erythrocytes, RNases, DNases, salts, etc. have to be removed because they tend to interfere with the microfluidic operations, downstream applications and subsequent analysis. These processes are highly dependent on the nature of the sample and are not necessarily small in scale (The WHO recommends the QIAmp Viral RNA Mini Kit (Qiagen) for RNA isolation5, which requires 140 μl raw sample as starting material). However, the design of an interface that allows the preconditioning of complex, real-world samples and the handling of RT-PCR volumes in the range of 1 pl–50 μl remains elusive17.
Conventional microfluidic systems are based on a continuous flow of liquids in silicon, glass or polymer microchannels equipped with external pumps, tubes, filters, mixers, etc.18. A new application, or the optimization of the system for a given application, usually requires another design that takes several days to fabricate. Droplet-based microfluidic architectures offer more flexibility—in particular, the ability to independently control each droplet, thus enabling complex (bio)chemical tasks to be processed in a fashion similar to that of a traditional biological laboratory19. In this context, the reconfiguration and scaling up of the system requires only rearranging the droplets and varying their volumes.
Popular techniques for actuating droplets on a planar surface rely on electrowetting20, dielectrophoresis21, acoustic waves22 or (electro) magnetic forces in combination with superparamagnetic particles23. By comparison, (electro)magnetic actuation has several unique properties. For example, the superparamagnetic particles can be remotely manipulated by a permanent magnet or an electromagnet located off-chip, allowing users to run dedicated tests in single-use disposables at low cost. Because the magnetic interaction is unaffected by surface charges, pH, ionic strength or temperature, it is compatible with a wide range of substrate materials and (bio)chemical processes. In addition, most prominent features of a bioassay, such as extraction, transport, mixing and detection, can be realized by exploiting superparamagnetic particles and their magnetic properties24.
Starting from a throat swab sample, we show how HPAI H5N1 can be detected by using magnetic forces to manipulate a free droplet containing superparamagnetic particles. The novelty of the method lies in the way that the droplet itself becomes a pump, valve, mixer, solid-phase extractor and real-time thermocycler. Interfacing the RNA sample preparation with real-time RT-PCR (RRT-PCR) makes the bioassay superior to tests currently marketed. (At present, the following RT-PCR kits for identifying H5N1 are commercially available: LightMix Kit for Influenza virus A H5, N1, M2 and H5N1 (TIB MOLBIOL/Roche), Avian Flu H5N1 RT-PCR Kit (1-step) (Veredus Laboratories), artus Influenza H5 LC RT-PCR Kit (Qiagen), and TaqMan Influenza A/H5 Kit v1.0 (Applied Biosystems). Including sample preparation, these kits claim an overall run time of around 4 h and rely on 20–50 μl RT-PCR mixture.)
Results
Droplet actuation
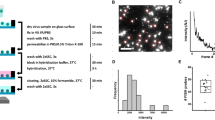
In our approach, a 100-nl droplet spontaneously forms on a perfluorinated glass or polymer chip after an aqueous suspension of surface-functionalized superparamagnetic particles is emulsified in an immiscible liquid (Fig. 1a). Encapsulating the droplet in mineral oil both reduces the friction between the aqueous droplet and the solid surface and prevents the evaporation of the aqueous phase. The volume ratio of the aqueous phase and the mineral oil is 100:1 for biochemical processes at room temperature and 1:5 for processes that require temperatures of up to 100 °C. Similar to a biphasic segmented flow in microchannels, the aqueous phase glides on a thin film of mineral oil25. This thin film is essential for handling highly viscous body fluids, such as a feces suspension, saliva or whole blood. Aside from their role as force mediators for manipulating the droplet in a magnetic field (Fig. 1b–g), the superparamagnetic particles serve temporarily as a solid phase for biochemical processes (Fig. 1h and Supplementary Fig. 1 online). Pulling the superparamagnetic particles out of the aqueous droplet during solid-phase extraction (SPE) requires raising their concentration by 1,000–2,500% to overcome the resistance imposed by the surface tension of the aqueous droplet. Owing to an inherent property of magnetic forces, the actuation of the droplet is distant from the surface, making it possible to place the droplet on a chip, run a dedicated test and then dispose of the chip. To perform temperature-controlled biochemical processes in real time within a droplet, the chip is placed on a microfabricated heater26 with an integrated optical detection system27.
(a) The droplet self-assembles on a perfluorinated surface after being laid as an aqueous suspension of superparamagnetic particles over mineral oil. (b–g) A permanent magnet is used to move (b), merge (c), mix (d) and split (e–g) droplets. Every droplet and/or droplet manipulation represents an item, equipment or task in a biological laboratory. For example, the sequence—merge, mix and split—emulates a two-dimensional SPE. The dead volume is close to zero and buffers are easily exchangeable. Silica-coated superparamagnetic particles enable the specific isolation, purification and preconcentration of nucleic acids. (h) After SPE, the immobilized viral RNA is magnetically pulled out of the raw sample solution, washed four times to remove residual contaminants, and desorbed into an RT-PCR solution positioned on top of a miniaturized real-time thermocycler (see also Supplementary Fig. 1). The mineral oil prevents the aqueous phase from evaporating at temperatures of up to 150 °C. G, perfluorinated glass substrate; M, permanent magnet; Su, superparamagnetic particles; B, aqueous buffer; I, thin film of immiscible liquid; N, nucleocapsid containing HPAI H5N1 RNA; L, lysis buffer; Sa, raw sample solution; W1 and W2, washing solutions; R, RT-PCR mixture; T, miniaturized thermocycler (in red).
The shrinking of dimensions leads to increased surface-to-volume ratios, which can cause biofouling induced by the nonspecific adsorption of various components of the bioassay. This will not occur with a free aqueous droplet positioned on a hydrophobic surface, where only a relatively small fraction of its surface is in contact with the substrate. Notably, a miniaturized thermocycler composed of a 500-nl aqueous droplet has a smaller surface-to-volume ratio (0.7 mm−1) than 50 μl of water in a 200-μl PCR tube (1.5 mm−1). In this context, the microfluidic system behaves like a macroscopic one and we do not undertake any additional measures to prevent biofouling.
SPE of RNA
The ability of the droplet to specifically retain or release the RNA after SPE of the throat swab sample is key for performing the test sequentially, because only the surface-bound material is passed on to the RT-PCR. After washing away the contaminants, there are two options for desorbing the RNA from the superparamagnetic particles: decreasing the ionic strength of the solution by eluting in water, or increasing the pH value and/or temperature of the solution28. Elution first into water and then into the RT-PCR solution would involve an additional step. Because the RT is usually carried out at a pH above 8.4 and at temperatures above 50 °C, it is more convenient to directly release the immobilized RNA into the droplet containing the RT-PCR mixture at processing temperature. However, the procedures show no difference with respect to their cycle threshold (CT) in the subsequent RRT-PCR that targets the hemagglutinin (HA) segment of HPAI H5N1 (data not shown).
The droplet-based SPE recovery of 1–50 RNA copies from the throat swab sample is a linear curve and shows an optimal PCR efficiency (Fig. 2), indicating that RNA is quantitatively reisolated and of high purity. The high yield and purity is attributable to the fact that there is no loss of material due to nonspecific adsorption to the substrate surface, because the RNA remains adsorbed to the superparamagnetic particles during the SPE. Potential contaminants, which could interfere with the PCR, remain in the Lysis/Binding Solution droplet (see Fig. 1). In addition, the surface-bound RNA is washed four times with 10 μl of washing solution. With a 1:100 volume ratio of the superparamagnetic particles to washing solution, the remaining impurities are diluted 100 million—that is, (1/100)4—times.
The CT values of seven separate experiments were plotted versus the log of the number of HPAI H5N1 RNA copies. The solid black circles are the CT values obtained after the droplet-based SPE; the solid red line is a linear regression fit (R2 = 0.971) of the CT values; the solid blue lines denote the upper and lower 95% confidence limits. The PCR efficiency (E) was 99%, E = 10(−1/slope)−1, where 'slope' is the slope of the linear regression fit. 0.78–50 in vitro transcribed HPAI H5N1 RNA copies were used in 50 μl RT-PCR master mixture. The RRT-PCR was performed off-chip using a DNA Engine Opticon 2 thermocycler (MJ Research).
The extraction of low copy numbers of RNA from a 100-μl raw sample volume with a 100-nl suspension of superparamagnetic particles corresponds to a preconcentration by 50,000%. This cannot be rivaled by any of the commercially available kits for the isolation of nucleic acids (as mentioned above), which only use a fraction of the eluted material for the succeeding RRT-PCR (see Supplementary Fig. 2 online).
RRT-PCR
After SPE, the RNA is desorbed from the superparamagnetic particles into 0.5–3 μl RT-PCR mixture at pH 8.4–8.7 and 50–61 °C over a period of 3–8 min. Depending on the type of RT-PCR kit that is used, the superparamagnetic particles are either removed or left in the RT-PCR mixture before the hot-start activation of the DNA polymerase.
Faster thermocycles are preferable for reducing the thermal burden on the DNA polymerase. To minimize the RT-PCR run time, each thermocycling step is shortened by decreasing the holding time and/or increasing the temperature, but this is done without compromising the PCR product specificity, yield and efficiency. Because we continuously monitor the fluorescence signal and the temperature within the droplet during the PCR, the thermocycling conditions can be optimized over the course of one experiment (Fig. 3). Shorter thermocycles are preferable for reducing the thermal burden on the DNA polymerase. Rather moderate heating and cooling rates of +11.5 °C/s and −5.6 °C/s, respectively, are a compromise between the low total mass of the thermocycler (36 mg) and the low heat conductivity of the disposable chip material. However, using a disposable chip avoids cross-contamination and keeps operational costs low. Although the passive cooling makes up 27% of the overall process time, we abstain from active cooling to reduce the overall power consumption. The overall RT-PCR run time for an experiment that uses a SYBR Green–based format with an antibody-modified hot-start polymerase is 21 min 40 s. Run times can be further shortened by using a two-step, rather than a three-step, thermocycling protocol and by reducing the difference between consecutive temperatures.
Only after the fluorescence signal reaches a plateau for a particular step is the temperature changed to the next level. After optimization, the time per thermocycle, including ramping, is 22 s. However, placing a disposable chip on top of the micromachined thermocycler more than outweighs the rather moderate ramping rates. The solid black line represents the temperature of the heater; the solid blue line shows the fluorescence intensity within the droplet. AU, arbitrary units.
Melting-curve analysis (Supplementary Fig. 3 online) and capillary electrophoresis (Supplementary Fig. 4 online) are used to verify the PCR product specificity and yield. Even after 50 PCR cycles, no nonspecifically amplified PCR products are observed. Depending on the RT-PCR volume, the PCR product yield is around 1–20 ng DNA per μl. The PCR efficiency over a dynamic range of six orders of magnitude is 93% (Fig. 4), whereby the correlation coefficient of 0.999 indicates the linearity of the linear regression fit. If the RT-PCR is carried out in 50-μl volumes in a conventional thermocycler, the PCR efficiency is close to 100% (data not shown). We hypothesize that the PCR efficiency in our system is affected by the evaporation of water from the RT-PCR buffer at elevated temperatures, which causes the relative concentration of the RT-PCR components to deviate from their optimal values. But because we only use an RT-PCR volume of 500 nl, the cost of a reaction is 2,000–5,000% lower than that of commercially available H5N1 tests, which rely on 20–50 μl buffer per RT-PCR cycle.
The CT values of seven separate experiments were plotted versus the log of the number of HPAI H5N1 cDNA copies. The solid black circles are CT values; the solid red line is a linear regression fit (R2 = 0.999) of the CT values; the solid blue lines denote the upper and lower 95% confidence limits. The PCR efficiency (E) was 93%, E = 10(−1/slope)−1. 12–1.2 × 107 copies of HPAI H5N1 cDNA were used in 500 nl RT-PCR mixture.
Discussion
With the exception of the concentration of the superparamagnetic particles, we follow established benchtop protocols for all single steps within the experiment. However, owing to the short diffusion distance between the surface of the superparamagnetic particles and the RNA during the SPE, as well as to the fast mass and heat transfer typical for a microsystem, the entire procedure is completed in less than 28 min (Fig. 5). This is about 440% faster than current H5N1 tests on the market that take around 4 h.
Starting with a raw throat swab sample, it took less than 28 min to complete the fully automated process. The single process times were: lysis/SPE, 300 s; four washes, 40 s; RT, 180 s; hot start, 30 s; PCR including melting curve analysis, 1,100 s. The solid black line represents the RNA sample, the solid red line is the no template control and the solid gray line denotes the threshold value. AU, arbitrary units. 500 in vitro–transcribed HPAI H5N1 RNA copies were used as input.
Point-of-care tests in low-resource settings demand low-cost, easy-to-use hand-held units that are ideally composed of an instrument and a disposable component. The basic design of our prototype instrument for the most part relies on that of a CD-ROM drive (Supplementary Fig. 5 online), making use of its power supply, its spindle stepper motor for microfluidic actuation and its optics for fluorescence detection. At present, we are working on the implementation of a second optical channel for an internal control that allows the user to check on the SPE of the RNA as well as for possible PCR inhibition. Other current experiments target transport- and storage-related issues by using anhydrous bioassay components that are stable at ambient temperatures.
In conclusion, we have demonstrated a fully integrated, droplet-based microfluidic system for detecting HPAI H5N1 directly from a throat swab sample within 28 min. The RNA was automatically isolated, purified and preconcentrated by 50,000% using a droplet containing superparamagnetic particles and was then subjected to 50 cycles of RRT-PCR. Aside from its being interfaced with sample preparation, the assay was as sensitive as, and 440% faster and 2,000–5,000% more economical than, other available tests. This strategy not only is applicable for this particular assay, but could easily be adapted for other infectious diseases, such as SARS, HIV and hepatitis B, by extracting nucleic acids from other body fluids, such as blood, urine or saliva. Because an increasing number of magnetic particle–based biochemical kits are now commercially available for the processing of cells, RNA, DNA and proteins, we envision the microfluidic system described here to be an attractive diagnostic platform, especially for decentralized environmental, biological or medical testing.
Methods
Surface chemistry.
Teflon-like surfaces were obtained by spin-coating VFM glass CoverSlips (CellPath) with a 1% solution of Teflon AF 1600 (DuPont) in FC-40 Fluorinert (3M). The films had a thickness of ∼100 nm and the static contact angles with water and with mineral oil (Sigma) were 110 ± 2° and 70 ± 2°, respectively.
Thermal management.
The thermal management of our microfluidic device is described in detail elsewhere26. We wetted the microfabricated heater(s) with 100 nl of mineral oil to improve the thermal contact with the perfluorinated chip. Before use, every chip was sterilized on top of the microfluidic device at 130 °C for a few minutes.
Optical detection.
Fluorescence was detected by a BX-51 Research Microscope (Olympus) equipped with an X-Cite 120 fluorescence illumination system (EXFO Life Sciences), an ET-GFP filter set (Chroma), a 378 Series M Plan Apo long-distance 20 × objective (Mitutoyo), and a H7421-40 photon-counting module (Hamamatsu). We recorded the spectra continuously using a TDS 5054B digital phosphor oscilloscope (Tektronix). Alternatively, we used our miniaturized optical detection system27. Typically, the last 500 ms of the elongation step are used to acquire real-time data. If not stated otherwise, the raw data are used.
Microfluidics.
The microfluidic device was mounted on a ProScanII motorized stage system (Prior Scientific), which was moved relative to a permanent N30H neodymium iron boron disc magnet (ASSEMtech EUROPE) to manipulate the droplets containing superparamagnetic particles. We controlled the x and y movement of the stage with LabVIEW 8 software (National Instruments).
SPE of RNA.
We used the MagMAX-96 Viral RNA Isolation Kit (Ambion) to obtain viral RNA. Because the original superparamagnetic particles had low magnetization, which made them unsuitable for some of the microfluidic manipulations, they were replaced by MagPrep silica particles (Merck) at a concentration of 200–500 μg per μl.
Because we did not have a biosafety level 3 laboratory in which to work with the virus itself, in vitro transcribed HPAI H5N1 RNA from either the artus Influenza/H5 LC RT-PCR Kit (Qiagen) or The Genome Institute of Singapore and the cDNA generated from each of these respective sources was used instead. A throat swab sample was taken from one of the authors with the Viral CULTURETTE Collection and Transport System (BD Diagnostics).
For the SPE of the RNA, we added 100 nl of MagPrep silica particles to a droplet containing 24.5 μl of throat swab sample spiked with in vitro–transcribed HPAI H5N1 RNA, 63.7 μl Lysis/Binding Solution (Ambion), 6.9 μl water, and 10 μl Lysis/Binding Enhancer (Ambion), mixed for 10 s by pipetting and lysed for 5 min. Thereafter, the superparamagnetic particles were split from the sample droplet and washed successively for 10 s in two droplets containing 10 μl Washing Solution 1 (Ambion) and two droplets containing 10 μl Washing Solution 2 (Ambion). After the SPE, the RNA was desorbed from the superparamagnetic particles.
Depending on the RT-PCR kit, we used the following conditions (pH, reverse transcription temperature (°C), reaction time (s)): QuantiTect SYBR Green RT-PCR Kit (Qiagen) (8.7, 50, 480), SuperScript III Platinum SYBR Green One-Step qRT-PCR Kit (Invitrogen) (8.4, 60, 180) and LightCycler RNA Master SYBR Green I Kit (Roche) (8.5, 61, 240).
RRT-PCR.
We used the SuperScript III Platinum SYBR Green One-Step qRT-PCR mixture (Invitrogen) to monitor the RT-PCR in real time under the following conditions: reverse transcription at 60 °C for 180 s, initial activation at 95 °C for 20 s, followed by 50 cycles of denaturation at 95 °C for 3 s, annealing at 56 °C for 12 s and elongation at 72 °C for 7 s. To control the temperature within the droplet, the quantum yield of SYBR Green was recorded as a function of the temperature during the hot start phase. The PCR was followed by a melting curve analysis using the following conditions: 95 °C for 3 s, 56 °C for 12 s, and ramping from 45 °C to 95 °C at 1 °C/s.
The 100-nl droplet containing the purified surface-bound RNA was moved onto the preheated microfabricated heater, merged with a droplet containing 0.5 μl of the SuperScript III Platinum SYBR Green One-Step qRT-PCR mixture (Invitrogen) and 2.5 μl mineral oil, mixed for 10 s, and subjected to the RT-PCR.
We selected the primers based on the sequence of the HPAI A H5N1 virus A/chicken/Hubei/327/2004 (CKXF) strain, National Center for Biotechnology Information (NCBI) accession number AY684706, and aligned them to all H5N1 subtypes available in the NCBI Influenza Virus Resource as of October 2005.
The target was a 114-base-pair (bp) fragment of the hemagglutinin (HA) segment of HPAI H5N1. We used 5′-CAA ACA GAT TAG TCC TTG CGA CTG-3′ (HA114U.v1) and 5′-CYT GCC ATC CTC CCT CTA TAA A-3′ (HA114L.v1) as forward and reverse primers, respectively.
For verifying the PCR product specificity and yield, we removed the magnetic particles and loaded the RT-PCR mixture into a DNA 500 LabChip Kit/Bioanalyzer 2100 (Agilent).
Note: Supplementary information is available on the Nature Medicine website.
Accession codes
References
Claas, E.C.J. Human influenza A H5N1 virus related to a highly pathogenic avian influenza virus. Lancet 351, 472–477 (1998).
Ungchusak, K. et al. Probable person-to-person transmission of avian influenza A (H5N1). N. Engl. J. Med. 352, 333–340 (2005).
Wilschut, J.C., McElhaney, J.E. & Palache, A.M. Influenza epidemics and pandemics. in Influenza (eds. Wilschut, J.C., McElhaney, J.E. & Palache, A.M.) 49–77 (Elsevier, Philadelphia, 2006).
Holmes, E.C., Taubenberger, J.K. & Grenfell, B.T. Heading off an influenza pandemic. Science 309, 989 (2005).
World Health Organization. WHO pandemic influenza draft protocol for rapid response and containment. 〈http://www.who.int/csr/disease/avian_influenza/guidelines/protocolfinal30_05_06a.pdf〉 (2007).
Ferguson, N.M. et al. Strategies for containing an emerging influenza pandemic in Southeast Asia. Nature 437, 209–214 (2005).
Longini, I.M., Jr. et al. Containing pandemic influenza at the source. Science 309, 1083–1087 (2005).
Ho, D. Is China prepared for microbial threats? Nature 435, 421–422 (2005).
Normile, D. Avian influenza. WHO proposes plan to stop pandemic in its tracks. Science 311, 315–316 (2006).
Abbott, A. & Pearson, H. Fear of human pandemic grows as bird flu sweeps through Asia. Nature 427, 472–473 (2004).
Enserink, M. Amid mayhem in Turkey, experts see new chances for research. Science 311, 314–315 (2006).
Yuen, K.Y. et al. Clinical features and rapid diagnosis of human disease associated with avian influenza A H5N1 virus. Lancet 351, 467–471 (1998).
Yager, P. et al. Microfluidic technologies for global public health. Nature 442, 412–418 (2006).
Whitesides, G.M. The origins and the future of microfluidics. Nature 442, 368–373 (2006).
de Mello, A.J. & Beard, N. Dealing with 'real' samples: sample pre-treatment in microfluidic systems. Lab Chip 3, 11N–19N (2003).
Auroux, P.-A., Koc, Y., deMello, A., Manz, A. & Day, J.R. Miniaturized nucleic acid analysis. Lab Chip 4, 534–546 (2004).
deMello, A.J. Control and detection of chemical reactions in microfluidic systems. Nature 442, 394–402 (2006).
Daw, R. & Finkelstein, J. Lab on a chip (Nature Insight). Nature 442, 367–412 (2006).
Mukhopadhyay, R. Diving into droplets. Anal. Chem. 78, 1401–1404 (2006).
Srinivasan, V., Pamula, V.K. & Fair, R.B. Droplet-based microfluidic lab-on-a-chip for glucose detection. Anal. Chim. Acta 507, 145–150 (2004).
Gascoyne, P.R. et al. Dielectrophoresis-based programmable fluidic processors. Lab Chip 4, 299–309 (2004).
Guttenberg, Z. et al. Planar chip device for PCR and hybridization with surface acoustic wave pump. Lab Chip 5, 308–317 (2005).
Lehmann, U., Vandevyver, C., Parashar, V.K. & Gijs, M.A. Droplet-based DNA purification in a magnetic lab-on-a-chip. Angew. Chem. Int. Ed. Engl. 45, 3062–3067 (2006).
Pamme, N. Magnetism and microfluidics. Lab Chip 6, 24–38 (2006).
Song, H., Chen, D.L. & Ismagilov, R.F. Reactions in droplets in microfluidic channels. Angew. Chem. Int. Edn Engl. 45, 7336–7356 (2006).
Neuzil, P., Zhang, C., Pipper, J., Oh, S. & Zhuo, L. Ultrafast miniaturized real-time PCR: 40 cycles in less than six minutes. Nucleic Acids Res. 34, e77 (2006).
Novak, L., Neuzil, P., Pipper, J., Zhang, Y. & Lee, S. An integrated fluorescence detection system for lab-on-a-chip applications. Lab Chip 7, 27–29 (2007).
Melzak, K.A., Sherwood, C.S., Turner, R.F.B. & Haynes, C.A. Driving forces for DNA adsorption to silica in perchlorate solutions. J. Colloid Interface Sci. 181, 635–644 (1996).
Acknowledgements
We thank A. Goh for reading the manuscript, K.S. Chee for taking the photographs and T.-M. Hsieh for assisting in software programming. This work was funded by the Singapore Institute of Bioengineering and Nanotechnology (IBN), the Singapore Biomedical Research Council (BMRC), and the Singapore Agency for Science, Technology and Research (A*STAR).
Author information
Authors and Affiliations
Contributions
J.P. supervised the project, conceived the experiments and wrote the manuscript. J.P. and Y.Z. performed the experiments and the data analyses. M.I. and L.F.-P.N. designed the PCR primers. P.N. fabricated the PCR chip. P.N. and L.N. designed the miniaturized detector. L.N. programmed the miniaturized thermocycler.
Corresponding author
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–5 (PDF 432 kb)
Rights and permissions
About this article
Cite this article
Pipper, J., Inoue, M., Ng, LP. et al. Catching bird flu in a droplet. Nat Med 13, 1259–1263 (2007). https://doi.org/10.1038/nm1634
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nm1634
This article is cited by
-
DropLab: an automated magnetic digital microfluidic platform for sample-to-answer point-of-care testing—development and application to quantitative immunodiagnostics
Microsystems & Nanoengineering (2023)
-
Magnetic Janus origami robot for cross-scale droplet omni-manipulation
Nature Communications (2023)
-
Aging Hearts in a Hotter, More Turbulent World: The Impacts of Climate Change on the Cardiovascular Health of Older Adults
Current Cardiology Reports (2022)
-
A 3D-printed magnetic digital microfluidic diagnostic platform for rapid colorimetric sensing of carbapenemase-producing Enterobacteriaceae
Microsystems & Nanoengineering (2021)
-
COVID-19 fear and volatility index movements: empirical insights from ASEAN stock markets
Environmental Science and Pollution Research (2021)