Abstract
The signaling adaptor MAVS forms prion-like aggregates to activate an innate antiviral immune response after viral infection. However, the molecular mechanisms that regulate MAVS aggregation are poorly understood. Here we identified TRIM31, an E3 ubiquitin ligase of the TRIM family of proteins, as a regulator of MAVS aggregation. TRIM31 was recruited to mitochondria after viral infection and specifically regulated antiviral signaling mediated by RLR pattern-recognition receptors. TRIM31-deficient mice were more susceptible to infection with RNA virus than were wild-type mice. TRIM31 interacted with MAVS and catalyzed the Lys63 (K63)-linked polyubiquitination of Lys10, Lys311 and Lys461 on MAVS. This modification promoted the formation of prion-like aggregates of MAVS after viral infection. Our findings reveal new insights in the molecular regulation of MAVS aggregation and the cellular antiviral response through TRIM31-mediated K63-linked polyubiquitination of MAVS.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout







Similar content being viewed by others
References
Kawai, T. & Akira, S. The role of pattern-recognition receptors in innate immunity: update on Toll-like receptors. Nat. Immunol. 11, 373–384 (2010).
Cao, X. Self-regulation and cross-regulation of pattern-recognition receptor signaling in health and disease. Nat. Rev. Immunol. 16, 35–50 (2016).
Wu, J. & Chen, Z.J. Innate immune sensing and signaling of cytosolic nucleic acids. Annu. Rev. Immunol. 32, 461–488 (2014).
Kato, H., Takahasi, K. & Fujita, T. RIG-I-like receptors: cytoplasmic sensors for non-self RNA. Immunol. Rev. 243, 91–98 (2011).
Unterholzner, L. et al. IFI16 is an innate immune sensor for intracellular DNA. Nat. Immunol. 11, 997–1004 (2010).
Sun, L., Wu, J., Du, F., Chen, X. & Chen, Z.J. Cyclic GMP–AMP synthase is a cytosolic DNA sensor that activates the type I interferon pathway. Science 339, 786–791 (2013).
Takeuchi, O. & Akira, S. Pattern-recognition receptors and inflammation. Cell 140, 805–820 (2010).
Yoneyama, M. & Fujita, T. Structural mechanism of RNA recognition by the RIG-I-like receptors. Immunity 29, 178–181 (2008).
Meylan, E. et al. Cardif is an adaptor protein in the RIG-I antiviral pathway and is targeted by hepatitis C virus. Nature 437, 1167–1172 (2005).
Seth, R.B., Sun, L., Ea, C.K. & Chen, Z.J. Identification and characterization of MAVS, a mitochondrial antiviral signaling protein that activates NF-κB and IRF3. Cell 122, 669–682 (2005).
Xu, L.G. et al. VISA is an adapter protein required for virus-triggered IFN-β signaling. Mol. Cell 19, 727–740 (2005).
Kawai, T. et al. IPS-1, an adaptor triggering RIG-I- and Mda5-mediated type I interferon induction. Nat. Immunol. 6, 981–988 (2005).
Hou, F. et al. MAVS forms functional prion-like aggregates to activate and propagate antiviral innate immune response. Cell 146, 448–461 (2011).
Peisley, A., Wu, B., Xu, H., Chen, Z.J. & Hur, S. Structural basis for ubiquitin-mediated antiviral signal activation by RIG-I. Nature 509, 110–114 (2014).
Moore, C.B. et al. NLRX1 is a regulator of mitochondrial antiviral immunity. Nature 451, 573–577 (2008).
Yoo, Y.S. et al. The mitochondrial ubiquitin ligase MARCH5 resolves MAVS aggregates during antiviral signaling. Nat. Commun. 6, 7910 (2015).
Sugiura, T. & Miyamoto, K. Characterization of TRIM31, upregulated in gastric adenocarcinoma, as a novel RBCC protein. J. Cell. Biochem. 105, 1081–1091 (2008).
Shi, M. et al. TRIM30-α negatively regulates TLR-mediated NF-κB activation by targeting TAB2 and TAB3 for degradation. Nat. Immunol. 9, 369–377 (2008).
Wang, P., Zhao, W., Zhao, K., Zhang, L. & Gao, C. TRIM26 negatively regulates interferon-β production and antiviral response through polyubiquitination and degradation of nuclear IRF3. PLoS Pathog. 11, e1004726 (2015).
Juang, Y.T. et al. Primary activation of interferon A and interferon B gene transcription by interferon regulatory factor 3. Proc. Natl. Acad. Sci. USA 95, 9837–9842 (1998).
Sugiura, T. The cellular level of TRIM31, an RBCC protein overexpressed in gastric cancer, is regulated by multiple mechanisms including the ubiquitin–proteasome system. Cell Biol. Int. 35, 657–661 (2011).
Gack, M.U. et al. TRIM25 RING-finger E3 ubiquitin ligase is essential for RIG-I-mediated antiviral activity. Nature 446, 916–920 (2007).
Zeng, W. et al. Reconstitution of the RIG-I pathway reveals a signaling role of unanchored polyubiquitin chains in innate immunity. Cell 141, 315–330 (2010).
Nakhaei, P., Genin, P., Civas, A. & Hiscott, J. RIG-I-like receptors: sensing and responding to RNA virus infection. Semin. Immunol. 21, 215–222 (2009).
Kawai, T. & Akira, S. Toll-like receptor and RIG-I-like receptor signaling. Ann. NY Acad. Sci. 1143, 1–20 (2008).
Xia, Z.P. et al. Direct activation of protein kinases by unanchored polyubiquitin chains. Nature 461, 114–119 (2009).
Larabi, A. et al. Crystal structure and mechanism of activation of TANK-binding kinase 1. Cell Rep. 3, 734–746 (2013).
Heaton, S.M., Borg, N.A. & Dixit, V.M. Ubiquitin in the activation and attenuation of innate antiviral immunity. J. Exp. Med. 213, 1–13 (2016).
You, F. et al. PCBP2 mediates degradation of the adaptor MAVS via the HECT ubiquitin ligase AIP4. Nat. Immunol. 10, 1300–1308 (2009).
Zhou, X., You, F., Chen, H. & Jiang, Z. Poly(C)-binding protein 1 (PCBP1) mediates housekeeping degradation of mitochondrial antiviral signaling (MAVS). Cell Res. 22, 717–727 (2012).
Zhong, B. et al. The E3 ubiquitin ligase RNF5 targets virus-induced signaling adaptor for ubiquitination and degradation. J. Immunol. 184, 6249–6255 (2010).
Arimoto, K. et al. Negative regulation of the RIG-I signaling by the ubiquitin ligase RNF125. Proc. Natl. Acad. Sci. USA 104, 7500–7505 (2007).
Wang, Y., Tong, X. & Ye, X. Ndfip1 negatively regulates RIG-I-dependent immune signaling by enhancing E3 ligase Smurf1-mediated MAVS degradation. J. Immunol. 189, 5304–5313 (2012).
Pan, Y. et al. Smurf2 negatively modulates RIG-I-dependent antiviral response by targeting VISA/MAVS for ubiquitination and degradation. J. Immunol. 192, 4758–4764 (2014).
Castanier, C. et al. MAVS ubiquitination by the E3 ligase TRIM25 and degradation by the proteasome is involved in type I interferon production after activation of the antiviral RIG-I-like receptors. BMC Biol. 10, 44 (2012).
Mori, M. et al. Identification of Ser386 of interferon regulatory factor 3 as critical target for inducible phosphorylation that determines activation. J. Biol. Chem. 279, 9698–9702 (2004).
Acknowledgements
We thank Z. Jiang (Peking University) for Mavs−/− MEFs; D. Guo (Wuhan University) for Ddx58−/− MEFs; K.A. Fitzgerald (University of Massachusetts Medical School) for the human IFNB1 promoter reporter and its mutated reporter; and B. Sun and F. Hou (Shanghai Institutes for Biological Sciences) for the expression plasmids for TRIM30-α and MAVS(M56R). Supported by the Natural Science Foundation of China (81525012, 81273219 and 81471538 to C.G.) and the Specialized Research Fund for the Doctoral Program of Higher Education of China (20130131130010 to C.G.).
Author information
Authors and Affiliations
Contributions
C.G. designed and supervised the research; B.L. executed the biochemical, cell biological and in vitro experiments; M.Z. performed the animal experiments, viral infections, and experiments with mutant MAVS and TRIM31; H.C., H.Z., H.W., G.S., P.W., K.Z., J.H., X.W. and L.Z. contributed reagents, analytical tools and discussions; B.L. prepared figures; and B.L and C.G. analyzed the data and wrote the paper.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Integrated supplementary information
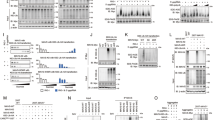
Supplementary Figure 1 TRIM31 is recruited to mitochondria after infection with SeV.
(a) Confocal microscopy of TRIM31-GFP transfected into HEK293T cells for 24 h followed with SeV infection for 6 h. MitoTracker (red) were used to label mitochondria. Scale bar 10 μm. (b) Immunoblot of TRIM31 protein and TOMM20 protein in the crude mitochondrial extracts (upper) and TRIM31 protein and β-actin in the whole cell lysate (lower) prepared from peritoneal macrophages infected with SeV for indicated times. (c) Immunoblot of TRIM31 protein in mouse peritoneal macrophages infected with SeV for indicated times. (d) qPCR analysis of Trim31, Trim26, Trim30α, Ifnb1 expression in primary peritoneal macrophages transfected with mice TRIM31 siRNA, TRIM26 siRNA, TRIM30α siRNA or control siRNA for 48 h, followed with SeV infection for indicated times. mRNA results are presented relative to those of untreated cells transfected with control siRNA. (e) qPCR analysis of TRIM31 and IFNB1 mRNA in THP-1 cells transfected with human TRIM31 siRNA or control siRNA for 48 h, followed with SeV infection for indicated times. mRNA results are presented as in d. (f) qPCR analysis of IFNB1 mRNA in Hela cells transfected with Flag-TRIM31 expression plasmid or control vector for 24 h, followed with SeV infection or transfection with poly(I:C) for the indicated times. mRNA results are presented relative to those of untreated cells transfected with control plasmid. The data are representative of 3 independent experiments with three biological replicates (means ± S.D. in d-f). *p < 0.01. (two-tailed Student’s t-test).
Supplementary Figure 2 TRIM31 deficiency impairs RNA-virus-induced production of type I interferon and antiviral responses.
(a) Schematic diagram of the deletions of the Trim31 deficient mice generated by transcription activation-like effector nuclease (TALEN) technology. The deletions were confirmed by DNA sequencing analysis of genomic DNA isolated from Trim31+/+ and Trim31–/– mice tails. (b) Immunoblot of TRIM31 protein in Trim31+/+ and Trim31–/– peritoneal macrophages. (c) qPCR analysis of Ifnb1, Ccl5 and Cxcl10 mRNA in the macrophages prepared form Trim31–/– (G) mice or Trim31–/– (5) mice infected with SeV for indicated times. mRNA results are presented relative to those of untreated wild-type cells. (d,e) qPCR analysis of Ifnb1, Ccl5 and Cxcl10 mRNA in bone marrow-derived macrophages (BMDMs) (d) or MEFs (e) prepared from Trim31+/+ and Trim31–/– mice followed with infection of SeV and HSV-1 or stimulation of 5′ppp-RNA, LPS and poly(I:C) for indicated times. mRNA results are presented as in c. The data are representative of 3 independent experiments with three biological replicates (means ± S.D. in c-e). *p < 0.05 and **p < 0.01. (two-tailed Student’s t-test).
Supplementary Figure 3 Trim31-deficient mice are not impaired in their response to infection with a DNA virus.
(a) qPCR analyses of Ifnb1 mRNA in brains and ELISA analysis of IFN-β production in sera of Trim31+/+ and Trim31–/– mice intraperitoneally infected with HSV-1 (2×107 PFU/mouse)for 24 h. mRNA results are presented relative to those of untreated wild-type cells. (b) qPCR analysis of HSV-1 genomic DNA and plaque assays of HSV-1 titer in brains of Trim31+/+ and Trim31–/– mice as treated in (a). (c) Survival of Trim31+/+ and Trim31–/– mice (n=7) after intraperitoneally injection of HSV-1 (1×108 pfu/mouse). The data are representative of 3 independent experiments with three biological replicates (means ± S.D. in a, b). *p < 0.01 (two-tailed Student’s t-test(a,b) or two-way analysis of variance (ANOVA) (c)).
Supplementary Figure 4 TRIM31 targets MAVS.
(a) Luciferase activity in HEK293T cells transfected with IFNB1 luciferase reporter and expression plasmids for RIG-IN, MDA5, MAVS, TBK1, IKKɛ and IRF3 5D together with Flag-TRIM31 expression plasmid for 24 h. (b) Luciferase activity in HEK293T cells transfected with control siRNA or TRIM31 siRNA and IFNB1 luciferase reporter or ISRE luciferase reporter together with expression plasmids for RIG-IN, MAVS and TBK1 for 24 h. (c) Coimmunoprecipitation analysis of Flag-TRIM31 interaction with Myc-IRF3 or Myc-STING in HEK293T cells. The data are representative of 3 independent experiments with three biological replicates (means ± S.D. in a, b). *p < 0.01. (two-tailed Student’s t-test).
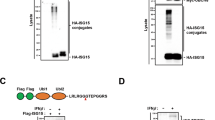
Supplementary Figure 5 TRIM31 catalyzes K63-linked polyubiquitination of MAVS.
(a) Coimmunoprecipitation analysis of RIG-I ubiquitination in HEK293T cells transfected with Myc-RIG-I and HA-Ubiquitin together with control plasmid, Flag-TRIM31 plasmid or Flag-TRIM25 plasmid. (b) Luciferase activity in HEK293T cells transfected with IFNB1 luciferase reporter or ISRE luciferase reporter together with Flag-TRIM31-WT plasmid or Flag-TRIM31-C53/56A plasmid for 24 h followed with SeV infection for indicated times. (c) qPCR analysis of IFNB1 mRNA and VSV RNA in HEK293T cells transfected with control plasmid, Flag-TRIM31-WT plasmid or Flag-TRIM31-C53/56A plasmid for 24 h followed with VSV infection (MOI, 0.1) for indicated times. mRNA results are presented relative to those of untreated cells transfected with control plasmid. (d) Immunoblot of phosphorylated and total IRF3 protein in lysates of HEK293T cells transfected with Flag-TRIM31-WT or Flag-TRIM31-C53/56A plasmid for 24 h followed with SeV infection for indicated times. The data are representative of 3 independent experiments with three biological replicates (means ± S.D. in b, c). *p < 0.01. (two-tailed Student’s t-test).
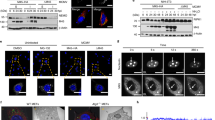
Supplementary Figure 6 TRIM31 promotes the formation of MAVS aggregates.
(a) SDD-AGE analysis of MAVS aggregation in Trim31+/+ and Trim31–/– MEFs infected with SeV for indicated times (upper). SDS-PAGE analysis of MAVS protein and TRIM31 protein in the whole cell lysates prepared above (lower). (b) SDD-AGE analysis of MAVS aggregation in HEK293T cells transfected with Flag-TRIM31 expression plasmid and Myc-MAVS for 24 h. (c) Immunoblot of TRIM31 protein in Trim31–/– MEFs reconstituted with mTRIM31-WT or mTRIM31-C52/55A. TRIM31+/+ MEFs were used as controls. qPCR analysis of Ifnb1, Ccl5 and Cxcl10 mRNA in TRIM31+/+ or Trim31–/– MEFs reconstituted with mTRIM31-WT or mTRIM31-C52/55A followed with SeV infection for 12 h. mRNA results are presented relative to those of untreated wide-type cells transfected with control plasmid. (d) qPCR analysis of Ifnb1 mRNA in Mavs–/– MEFs transfected with MAVS expression plasmid or control plasmid for 24 h followed with SeV infection for 12 h. mRNA results are presented relative to those of untreated cells transfected with control plasmid. (e) Luciferase activity in Mavs–/– MEFs transfected with IFN-β or ISRE luciferase reporter and expression plasmids for MAVS-WT, MAVS-K10R, MAVS-K10/311R, MAVS-K10/311/461R along with Flag-TRIM31 expression plasmid for 24 h followed with SeV infection for 12 h. (f) qPCR analysis of Ifnb1 mRNA and SDD-AGE analysis of MAVS aggregation in Mavs–/– MEFs transfected with MAVS-WT, MAVS-K10R, MAVS-K311R and MAVS-K461R along with Flag-TRIM31 expression plasmid or control plasmid for 24 h followed with SeV infection for 12 h. (g) qPCR analysis of Ifnb1 mRNA and SDD-AGE analysis of MAVS aggregation in Mavs–/– MEFs transfected with MAVS-WT or MAVS-W56R along with Flag-TRIM31 expression plasmid or control plasmid for 24 h followed with SeV infection for 6 h. mRNA results are presented as in d. The data are representative of 3 independent experiments with three biological replicates (means ± S.D. in c-g). *p < 0.01. (two-tailed Student’s t-test).
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–6 and Supplementary Tables 1 and 2 (PDF 1793 kb)
Rights and permissions
About this article
Cite this article
Liu, B., Zhang, M., Chu, H. et al. The ubiquitin E3 ligase TRIM31 promotes aggregation and activation of the signaling adaptor MAVS through Lys63-linked polyubiquitination. Nat Immunol 18, 214–224 (2017). https://doi.org/10.1038/ni.3641
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ni.3641
This article is cited by
-
O-GlcNAc of STING mediates antiviral innate immunity
Cell Communication and Signaling (2024)
-
OTUD5 promotes the inflammatory immune response by enhancing MyD88 oligomerization and Myddosome formation
Cell Death & Differentiation (2024)
-
The E3 ubiquitin ligase TRIM31 attenuates NLRP3 inflammasome activation in Helicobacter pylori-associated gastritis by regulating ROS and autophagy
Cell Communication and Signaling (2023)
-
MAVS-loaded unanchored Lys63-linked polyubiquitin chains activate the RIG-I-MAVS signaling cascade
Cellular & Molecular Immunology (2023)
-
MAVS deSUMOylation by SENP1 inhibits its aggregation and antagonizes IRF3 activation
Nature Structural & Molecular Biology (2023)