Abstract
The differentiation of helper T cells into effector subsets is critical to host protection. Transcription factors of the E-protein and Id families are important arbiters of T cell development, but their role in the differentiation of the TH1 and TFH subsets of helper T cells is not well understood. Here, TH1 cells showed more robust Id2 expression than that of TFH cells, and depletion of Id2 via RNA-mediated interference increased the frequency of TFH cells. Furthermore, TH1 differentiation was blocked by Id2 deficiency, which led to E-protein-dependent accumulation of effector cells with mixed characteristics during viral infection and severely impaired the generation of TH1 cells following infection with Toxoplasma gondii. The TFH cell–defining transcriptional repressor Bcl6 bound the Id2 locus, which provides a mechanism for the bimodal Id2 expression and reciprocal development of TH1 cells and TFH cells.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
Change history
02 June 2016
In the version of this article initially published online, the label above the far right plot in Figure 3g ('Blc6') was incorrect. The correct label is 'Bcl6'. The error has been corrected for the print, PDF and HTML versions of this article.
References
Zhu, J., Yamane, H. & Paul, W.E. Differentiation of effector CD4 T cell populations. Annu. Rev. Immunol. 28, 445–489 (2010).
Crotty, S. T follicular helper cell differentiation, function, and roles in disease. Immunity 41, 529–542 (2014).
Vahedi, G. et al. Helper T-cell identity and evolution of differential transcriptomes and epigenomes. Immunol. Rev. 252, 24–40 (2013).
Nurieva, R.I. et al. Bcl6 mediates the development of T follicular helper cells. Science 325, 1001–1005 (2009).
Yu, D. et al. The transcriptional repressor Bcl-6 directs T follicular helper cell lineage commitment. Immunity 31, 457–468 (2009).
Nakayamada, S. et al. Early Th1 cell differentiation is marked by a Tfh cell-like transition. Immunity 35, 919–931 (2011).
Oestreich, K.J., Mohn, S.E. & Weinmann, A.S. Molecular mechanisms that control the expression and activity of Bcl-6 in TH1 cells to regulate flexibility with a TFH-like gene profile. Nat. Immunol. 13, 405–411 (2012).
Johnston, R.J. et al. Bcl6 and Blimp-1 are reciprocal and antagonistic regulators of T follicular helper cell differentiation. Science 325, 1006–1010 (2009).
Cannarile, M.A. et al. Transcriptional regulator Id2 mediates CD8+ T cell immunity. Nat. Immunol. 7, 1317–1325 (2006).
de Pooter, R.F. & Kee, B.L. E proteins and the regulation of early lymphocyte development. Immunol. Rev. 238, 93–109 (2010).
D'Cruz, L.M., Stradner, M.H., Yang, C.Y. & Goldrath, A.W. E and Id proteins influence invariant NKT cell sublineage differentiation and proliferation. J. Immunol. 192, 2227–2236 (2014).
Jones-Mason, M.E. et al. E protein transcription factors are required for the development of CD4+ lineage T cells. Immunity 36, 348–361 (2012).
Eberl, G., Colonna, M., Di Santo, J.P. & McKenzie, A.N. Innate lymphoid cells. Innate lymphoid cells: a new paradigm in immunology. Science 348, aaa6566 (2015).
Yang, C.Y. et al. The transcriptional regulators Id2 and Id3 control the formation of distinct memory CD8+ T cell subsets. Nat. Immunol. 12, 1221–1229 (2011).
D'Cruz, L.M., Lind, K.C., Wu, B.B., Fujimoto, J.K. & Goldrath, A.W. Loss of E protein transcription factors E2A and HEB delays memory-precursor formation during the CD8+ T-cell immune response. Eur. J. Immunol. 42, 2031–2041 (2012).
Masson, F. et al. Id2-mediated inhibition of E2A represses memory CD8+ T cell differentiation. J. Immunol. 190, 4585–4594 (2013).
Ji, Y. et al. Repression of the DNA-binding inhibitor Id3 by Blimp-1 limits the formation of memory CD8+ T cells. Nat. Immunol. 12, 1230–1237 (2011).
Gao, P. et al. Dynamic changes in E-protein activity regulate T reg cell development. J. Exp. Med. 211, 2651–2668 (2014).
Maruyama, T. et al. Control of the differentiation of regulatory T cells and TH17 cells by the DNA-binding inhibitor Id3. Nat. Immunol. 12, 86–95 (2011).
Miyazaki, M. et al. Id2 and Id3 maintain the regulatory T cell pool to suppress inflammatory disease. Nat. Immunol. 15, 767–776 (2014).
Lin, Y.Y. et al. Transcriptional regulator Id2 is required for the CD4 T cell immune response in the development of experimental autoimmune encephalomyelitis. J. Immunol. 189, 1400–1405 (2012).
Liu, X. et al. Transcription factor achaete-scute homologue 2 initiates follicular T-helper-cell development. Nature 507, 513–518 (2014).
Choi, Y.S. et al. Bcl6 expressing follicular helper CD4 T cells are fate committed early and have the capacity to form memory. J. Immunol. 190, 4014–4026 (2013).
Miyazaki, M. et al. The opposing roles of the transcription factor E2A and its antagonist Id3 that orchestrate and enforce the naive fate of T cells. Nat. Immunol. 12, 992–1001 (2011).
Choi, Y.S. et al. ICOS receptor instructs T follicular helper cell versus effector cell differentiation via induction of the transcriptional repressor Bcl6. Immunity 34, 932–946 (2011).
Choi, Y.S. et al. LEF-1 and TCF-1 orchestrate TFH differentiation by regulating differentiation circuits upstream of the transcriptional repressor Bcl6. Nat. Immunol. 16, 980–990 (2015).
Nance, J.P. et al. Bcl6 middle domain repressor function is required for T follicular helper cell differentiation and utilizes the corepressor MTA3. Proc. Natl. Acad. Sci. USA 112, 13324–13329 (2015).
Niola, F. et al. Id proteins synchronize stemness and anchorage to the niche of neural stem cells. Nat. Cell Biol. 14, 477–487 (2012).
Johnston, R.J., Choi, Y.S., Diamond, J.A., Yang, J.A. & Crotty, S. STAT5 is a potent negative regulator of TFH cell differentiation. J. Exp. Med. 209, 243–250 (2012).
Poholek, A.C. et al. In vivo regulation of Bcl6 and T follicular helper cell development. J. Immunol. 185, 313–326 (2010).
Ray, J.P. et al. The interleukin-2-mTORc1 kinase axis defines the signaling, differentiation, and metabolism of T helper 1 and follicular B helper T cells. Immunity 43, 690–702 (2015).
Amir, A.D. et al. viSNE enables visualization of high dimensional single-cell data and reveals phenotypic heterogeneity of leukemia. Nat. Biotechnol. 31, 545–552 (2013).
Sher, A. et al. Induction and regulation of IL-12-dependent host resistance to Toxoplasma gondii. Immunol. Res. 27, 521–528 (2003).
Guo, Z. et al. Modeling Sjögren's syndrome with Id3 conditional knockout mice. Immunol. Lett. 135, 34–42 (2011).
Nance, J.P., Bélanger, S., Johnston, R.J., Takemori, T. & Crotty, S. Cutting edge: T follicular helper cell differentiation is defective in the absence of Bcl6 BTB repressor domain function. J. Immunol. 194, 5599–5603 (2015).
Pepper, M., Pagán, A.J., Igyártó, B.Z., Taylor, J.J. & Jenkins, M.K. Opposing signals from the Bcl6 transcription factor and the interleukin-2 receptor generate T helper 1 central and effector memory cells. Immunity 35, 583–595 (2011).
Hatzi, K. et al. BCL6 orchestrates Tfh cell differentiation via multiple distinct mechanisms. J. Exp. Med. 212, 539–553 (2015).
Xiao, R. et al. Identification and characterization of a cathepsin D homologue from lampreys (Lampetra japonica). Dev. Comp. Immunol. 49, 149–156 (2015).
Stone, E.L. et al. ICOS coreceptor signaling inactivates the transcription factor FOXO1 to promote Tfh cell differentiation. Immunity 42, 239–251 (2015).
Miyazaki, M. et al. The E-Id protein axis modulates the activities of the PI3K-AKT-mTORC1-Hif1a and c-myc/p19Arf pathways to suppress innate variant TFH cell development, thymocyte expansion, and lymphomagenesis. Genes Dev. 29, 409–425 (2015).
Kitano, M. et al. Bcl6 protein expression shapes pre-germinal center B cell dynamics and follicular helper T cell heterogeneity. Immunity 34, 961–972 (2011).
Hale, J.S. et al. Distinct memory CD4+ T cells with commitment to T follicular helper- and T helper 1-cell lineages are generated after acute viral infection. Immunity 38, 805–817 (2013).
Yusuf, I. et al. Germinal center T follicular helper cell IL-4 production is dependent on signaling lymphocytic activation molecule receptor (CD150). J. Immunol. 185, 190–202 (2010).
Lin, Y.C. et al. A global network of transcription factors, involving E2A, EBF1 and Foxo1, that orchestrates B cell fate. Nat. Immunol. 11, 635–643 (2010).
Wu, T. et al. TCF1 is required for the T follicular helper cell response to viral infection. Cell Rep. 12, 2099–2110 (2015).
Xu, L. et al. The transcription factor TCF-1 initiates the differentiation of TFH cells during acute viral infection. Nat. Immunol. 16, 991–999 (2015).
Kroenke, M.A. et al. Bcl6 and Maf cooperate to instruct human follicular helper CD4 T cell differentiation. J. Immunol. 188, 3734–3744 (2012).
Choi, Y.S., Eto, D., Yang, J.A., Lao, C. & Crotty, S. Cutting edge: STAT1 is required for IL-6-mediated Bcl6 induction for early follicular helper cell differentiation. J. Immunol. 190, 3049–3053 (2013).
Baumjohann, D., Okada, T. & Ansel, K.M. Cutting Edge: distinct waves of BCL6 expression during T follicular helper cell development. J. Immunol. 187, 2089–2092 (2011).
Oxenius, A., Bachmann, M.F., Zinkernagel, R.M. & Hengartner, H. Virus-specific MHC-class II-restricted TCR-transgenic mice: effects on humoral and cellular immune responses after viral infection. Eur. J. Immunol. 28, 390–400 (1998).
Kaji, T. et al. Distinct cellular pathways select germline-encoded and somatically mutated antibodies into immunological memory. J. Exp. Med. 209, 2079–2097 (2012).
Chen, R. et al. In vivo RNA interference screens identify regulators of antiviral CD4+ and CD8+ T cell differentiation. Immunity 41, 325–338 (2014).
Doedens, A.L. et al. Hypoxia-inducible factors enhance the effector responses of CD8+ T cells to persistent antigen. Nat. Immunol. 14, 1173–1182 (2013).
Ebert, A. et al. The distal VH gene cluster of the Igh locus contains distinct regulatory elements with Pax5 transcription factor-dependent activity in pro-B cells. Immunity 34, 175–187 (2011).
Acknowledgements
We thank the members of the Goldrath and Crotty laboratories for discussions; B. Yu for assistance with bioinformatics analysis; and I. Bilic and M. Busslinger for E2A Bio-ChiP-seq data. Supported by the US National Institutes of Health (1F31AG043222-01A1 to L.A.S., AI108651 to L.-F.L., AI067545 to A.W.G., AI109976 to A.W.G. and S.C.), the Fonds de la recherche Québec–Santé (Postdoctoral Training Award to S.B.), The Damon Runyon Cancer Research Foundation (Fraternal Order of Eagles Fellowship DRG-2069-11 to J.P.S.-B.) and the Leukemia and Lymphoma Society (K.D.O. and A.W.G.).
Author information
Authors and Affiliations
Contributions
L.A.S. and S.B. performed experiments; K.D.O., Su.C., J.P.S.-B., J.P.N., J.G., A.L. and L.-F.L. provided intellectual input and generated new reagents or performed experiments; and L.A.S., S.B., Sh.C. and A.W.G. conceived of the study, analyzed and interpreted data, and wrote the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Integrated supplementary information
Supplementary Figure 1 Id2 and Id3 define polyclonal TH1 and TFH cell subsets.
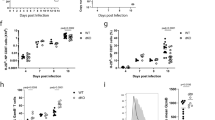
Id2YFP/+ (a) or Id3GFP/+ (b) mice were analyzed 7 days after LCMV infection. TH1 (SLAM+CXCR5− or CXCR5−PD-1−), TFH (SLAMloCXCR5+ or CXCR5+PD-1−) or GC TFH (CXCR5+PD-1+) differentiation for the indicated antigen-experienced (CD49d+CD11a+) CD4+ T cell populations was analyzed by flow cytometry and quantified. *p<0.01, **p<0.001 and ***p<0.0001 (two-tailed unpaired Student's t test). Data are representative of three experiments (a,b), each with n = 3 mice per group (mean ± s.e.m.).
Supplementary Figure 2 Knockdown of Id2 results in increased TFH differentiation.
SMARTA CD4+ T cells transduced with the indicated shRNAmir-RV were transferred into B6 SMARTA CD4+ T cells were transduced with the indicated shRNAmir-RV. (a) RNA was isolated from shRNAmir-RV+ CD4+ T cells and Id2 expression was determined by qRT-PCR. shRNAmir-RV+ CD4+T cells were transferred into B6 mice and analyzed 6 (b-e) or 3 (f-i) days after LCMV infection. (b,f) Quantitation of shRNA+ SMARTA CD4+ T cells. (c-e) GC TFH (CXCR5+PSGL-1−) cell development was analyzed by flow cytometry (c) and quantified as a fraction of SMARTA CD4+ T cells (d) or total splenocytes (e). (g-i) TFH (CXCR5+CD25−) and TH1 (CD25+CXCR5−) differentiation was analyzed by flow cytometry (g) and quantified as a fraction of SMARTA CD4+ T cells (h) or total splenocytes (i). (j,k) OT-II CD4+ T cells transduced with the indicated shRNAmir-RV were transferred into B6 mice and analyzed 11 days after footpad immunization with NP-OVA in alum. (j) TFH (CXCR5+PD-1+) and (k) GC TFH (CXCR5+Bcl6+) differentiation was analyzed by flow cytometry and quantified as a fraction of OT-II CD4+ T cells or total lymph node cells. *p<0.05, **p<0.01, ***p<0.001, ****p<0.0001 (two-tailed unpaired Student's t test). Data are pooled from two (a) experiments or representative of two (j,k), four (b-e) or five (f-i) independent experiments with n=6-14 mice per group (mean ± s.e.m.).
Supplementary Figure 3 Expression of Id proteins orchestrates CD4+ T cell differentiation.
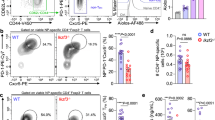
(a-c) Id2+/+ CD4-Cre+ (Id2+/+) or Id2fl/fl CD4-Cre+ (Id2−/−) SMARTA CD4+ T cells were transferred into B6 mice and analyzed 4 (a) and 7 (a-c) days after LCMV infection. (a) Flow cytometric analysis of CXCR5 and SLAM expression quantified as gMFI on days 4 and 7. (b) Flow cytometric analysis of granzyme B and TCF1 expression (left panels), quantification as a frequency of SMARTA CD4+ T cells (right panels). (c) Flow cytometric analysis of IFN-γ and T-bet expression in total SMARTA CD4+ T cells. gMFI of T-bet expression and total number of IFN-y+ SMARTA CD4+ T cells is shown. (d) Id2+/+ CD4-Cre+ and Id2fl/fl CD4-Cre+ mice were analyzed 7 days after LCMV infection, SLAMhiCXCR5−, SLAMmidCXCR5mid and SLAMloCXCR5+ cells were analyzed by flow cytometry (left panels) and quantified as a frequency of antigen-experienced (CD49d+CD11a+) CD4+ T cells (middle panel) or as total splenic numbers (right panel). (e) SMARTA CD4+ T cells transduced with the indicated shRNAmir-RV were transferred into B6 mice and analyzed 3 days after LCMV infection. Quantitation of Bcl6 expression in SMARTA TH1 (CXCR5−PD-1−) and TFH (CXCR5+PD-1+) cells is shown. (f) Id2+/+ CD4-Cre+ or Id2fl/fl CD4-Cre+ SMARTA CD4+ T cells were transferred into Bcl6fl/fl CD4-Cre+ mice and analyzed 8 days after LCMV infection. SLAMhiCXCR5−, SLAMmidCXCR5mid and SLAMloCXCR5+ cells were analyzed by flow cytometry (left panels) and quantified as a frequency of SMARTA CD4+ T cells (middle panel) or as total numbers (right panel) *p<0.05, **p<0.01, ***p<0.001, ****p<0.0001 (two-tailed unpaired Student's t test). Data are representative of one (e) or three (a-d,f) independent experiments with n=3-10 mice per group (mean ± s.e.m.).
Supplementary Figure 4 Expression of Id3 limits unregulated differentiation of TFH cells and GC TFH cells.
(a) Id3+/+ CD4-Cre+ (Id3+/+) or Id3fl/fl CD4-Cre+ (Id3−/−) SMARTA CD4+ T cells were transferred into B6 mice and analyzed 7 days after LCMV infection. (a) Flow cytometric analysis of CXCR5+Bcl6hi (top), SLAMhiCXCR5− (TH1) and SLAMloCXCR5+ (TFH) (middle); or PD-1−CXCR5− (TH1), PD-1−CXCR5+ (TFH) and PD-1+CXCR5+ (GC TFH) (bottom) populations and quantification as a frequency of SMARTA CD4+ T cells (right panels). (b) Id3+/+ CD4-Cre+ and Id3fl/fl CD4-Cre+ mice were analyzed 7 days after LCMV and PD-1−CXCR5− (TH1), PD-1−CXCR5+ (TFH) and PD-1+CXCR5+ (GC TFH) expression was analyzed by flow cytometry (left) and quantified as a frequency of antigen-specific (gp66-77) CD4+ T cells (right). (c-d) NIP CD4+ T cells transduced with the indicated RV were transferred into B6 mice and analyzed 6 days (c) or 3 days (d) after LCMV infection. (c) TFH (CXCR5+SLAMlo) differentiation was analyzed by flow cytometry and quantified as a frequency of NIP CD4+ T cells. (d) Early TFH (CXCR5+Bcl6+) differentiation was analyzed by flow cytometry and quantified as a frequency of NIP CD4+ T cells. *p<0.05, **p<0.01, ***p<0.001, ****p<0.0001 (two-tailed unpaired Student's t test). Data are pooled from two (c,d) experiments or representative of two (a,b) independent experiments with n=8-10 mice per group (mean ± s.e.m.).
Supplementary Figure 5 E proteins drive CXCR5 expression and inhibit the formation of TH1 cells.
(a) Id2+/+ CD4-Cre+ or Id2fl/fl CD4-Cre+ SMARTA CD4+ T cells were transduced with the indicated shRNAmir-RV and expanded in vitro for 4 days. Graph indicates relative mRNA expression of Tcf3 by dsRED+ SMARTA CD4+ T cells. (b) Gene expression of E proteins and related genes of interest in TH1 and TFH SMARTA at day 3 after acute LCMV infection measured by RNA-Seq. Cd8a and Cd19 are included as negative controls. Data are from ref. 32 (GSE67336). SMARTA (c) or NIP (d-k) CD4+ T cells transduced with the indicated RV were transferred into B6 mice and analyzed 3 days (c-f) or 6 days (g-k) after LCMV infection. (c) CXCR5 expression by SMARTA TH1 (CXCR5−Bcl6−) and TFH (CXCR5+Bcl6+) cells. (d,g) Quantitation of RV+ NIP CD4+ T cells. (e) CXCR5 expression by NIP TH1 (CXCR5−Bcl6−) and TFH (CXCR5+Bcl6+) cells. (f) Quantitation of Tbet expression by NIP TH1 (CD25+CXCR5−) and TFH (CXCR5+CD25−) cells. (h-j) TFH (CXCR5+SLAMlo) and TH1 (SLAM+CXCR5−) differentiation was analyzed by flow cytometry (h) and quantified as a fraction of NIP CD4+ T cells (i) or total splenocytes (j). (k) CXCR5 expression by NIP TH1 (SLAM+CXCR5 −) and TFH (CXCR5+SLAMlo) cells.. **p<0.01, ***p<0.0001 (two-tailed unpaired Student's t test). Data are pooled from two (i,j) experiments or are representative of two (a,c) or three (d-h,k) independent experiments with n=8-10 mice per group (mean ± s.e.m.).
Supplementary Figure 6 Bcl6 inhibits Id2 expression.
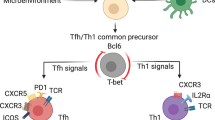
(a) Bcl6fl/fl CD4-Cre+ SMARTA CD4+ T cells transduced with the indicated vectors were transferred into B6 mice. (b) WT, Bcl6fl/WT CD4-Cre+ (Bcl6+/–), Bcl6fl/fl CD4-Cre+ (Bcl6−/−) SMARTA CD4+ T cells were transferred into B6 mice. Gates used to sort IL-2Rα+ and IL-2Rα− SMARTA CD4+ T cells 3 days after LCMV infection are indicated. (c) Sequences of the primers used in the study. (d) Model for the role of Id and E proteins in orchestrating CD4+ T cell differentiation. Data are representative of 2 independent experiments.
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–6 (PDF 1318 kb)
Rights and permissions
About this article
Cite this article
Shaw, L., Bélanger, S., Omilusik, K. et al. Id2 reinforces TH1 differentiation and inhibits E2A to repress TFH differentiation. Nat Immunol 17, 834–843 (2016). https://doi.org/10.1038/ni.3461
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ni.3461
This article is cited by
-
Spatiotemporal resolution of germinal center Tfh cell differentiation and divergence from central memory CD4+ T cell fate
Nature Communications (2023)
-
Synthetically glycosylated antigens for the antigen-specific suppression of established immune responses
Nature Biomedical Engineering (2023)
-
Pathogen-associated T follicular helper cell plasticity is critical in anti-viral immunity
Science China Life Sciences (2022)
-
Transcription tipping points for T follicular helper cell and T-helper 1 cell fate commitment
Cellular & Molecular Immunology (2021)
-
METTL3-dependent m6A modification programs T follicular helper cell differentiation
Nature Communications (2021)