Abstract
CD11b+ dendritic cells (DCs) seem to be specialized for presenting antigens via major histocompatibility (MHC) class II complexes to stimulate helper T cells, but the genetic and regulatory basis for this is not established. Conditional deletion of Irf4 resulted in loss of CD11b+ DCs, impaired formation of peptide–MHC class II complexes and defective priming of helper T cells but not of cytotoxic T lymphocyte (CTL) responses. Gene expression and chromatin immunoprecipitation followed by deep sequencing (ChIP-Seq) analyses delineated an IRF4-dependent regulatory module that programs enhanced MHC class II antigen presentation. Expression of the transcription factor IRF4 but not of IRF8 restored the ability of IRF4-deficient DCs to efficiently process and present antigen to MHC class II–restricted T cells and promote helper T cell responses. We propose that the evolutionary divergence of IRF4 and IRF8 facilitated the specialization of DC subsets for distinct modes of antigen presentation and priming of helper T cell versus CTL responses.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
Accession codes
References
Mellman, I. & Steinman, R.M. Dendritic cells: specialized and regulated antigen processing machines. Cell 106, 255–258 (2001).
Dudziak, D. et al. Differential antigen processing by dendritic cell subsets in vivo. Science 315, 107–111 (2007).
Satpathy, A.T., Wu, X., Albring, J.C. & Murphy, K.M. Re(de)fining the dendritic cell lineage. Nat. Immunol. 13, 1145–1154 (2012).
Edelson, B.T. et al. Peripheral CD103+ dendritic cells form a unified subset developmentally related to CD8α+ conventional dendritic cells. J. Exp. Med. 207, 823–836 (2010).
Hildner, K. et al. Batf3 deficiency reveals a critical role for CD8α+ dendritic cells in cytotoxic T cell immunity. Science 322, 1097–1100 (2008).
Suzuki, S. et al. Critical roles of interferon regulatory factor 4 in CD11bhighCD8α− dendritic cell development. Proc. Natl. Acad. Sci. USA 101, 8981–8986 (2004).
Tamura, T. et al. IFN regulatory factor-4 and -8 govern dendritic cell subset development and their functional diversity. J. Immunol. 174, 2573–2581 (2005).
Bajaña, S., Roach, K., Turner, S., Paul, J. & Kovats, S. IRF4 promotes cutaneous dendritic cell migration to lymph nodes during homeostasis and inflammation. J. Immunol. 189, 3368–3377 (2012).
Persson, E.K. et al. IRF4 transcription-factor-dependent CD103+CD11b+ dendritic cells drive mucosal T helper 17 cell differentiation. Immunity 38, 958–969 (2013).
Schlitzer, A. et al. IRF4 transcription factor-dependent CD11b+ dendritic cells in human and mouse control mucosal IL-17 cytokine responses. Immunity 38, 970–983 (2013).
Yamamoto, M. et al. Shared and distinct functions of the transcription factors IRF4 and IRF8 in myeloid cell development. PLoS ONE 6, e25812 (2011).
Klein, U. et al. Transcription factor IRF4 controls plasma cell differentiation and class-switch recombination. Nat. Immunol. 7, 773–782 (2006).
Caton, M.L., Smith-Raska, M.R. & Reizis, B. Notch-RBP-J signaling controls the homeostasis of CD8− dendritic cells in the spleen. J. Exp. Med. 204, 1653–1664 (2007).
Thomsen, A.R., Johansen, J., Marker, O. & Christensen, J.P. Exhaustion of CTL memory and recrudescence of viremia in lymphocytic choriomeningitis virus-infected MHC class II-deficient mice and B cell-deficient mice. J. Immunol. 157, 3074–3080 (1996).
Rudensky, A., Rath, S., Preston-Hurlburt, P., Murphy, D.B. & Janeway, C.A. Jr. On the complexity of self. Nature 353, 660–662 (1991).
Miller, J.C. et al. Deciphering the transcriptional network of the dendritic cell lineage. Nat. Immunol. 13, 888–899 (2012).
Heng, T.S. & Painter, M.W. The Immunological Genome Project: networks of gene expression in immune cells. Nat. Immunol. 9, 1091–1094 (2008).
Robbins, S.H. et al. Novel insights into the relationships between dendritic cell subsets in human and mouse revealed by genome-wide expression profiling. Genome Biol. 9, R17 (2008).
Amit, I. et al. Unbiased reconstruction of a mammalian transcriptional network mediating pathogen responses. Science 326, 257–263 (2009).
Meredith, M.M. et al. Zinc finger transcription factor zDC is a negative regulator required to prevent activation of classical dendritic cells in the steady state. J. Exp. Med. 209, 1583–1593 (2012).
Wu, L. et al. RelB is essential for the development of myeloid-related CD8α− dendritic cells but not of lymphoid-related CD8α+ dendritic cells. Immunity 9, 839–847 (1998).
Chan, Y.H. et al. Absence of the transcriptional repressor Blimp-1 in hematopoietic lineages reveals its role in dendritic cell homeostatic development and function. J. Immunol. 183, 7039–7046 (2009).
Muhlethaler-Mottet, A., Otten, L.A., Steimle, V. & Mach, B. Expression of MHC class II molecules in different cellular and functional compartments is controlled by differential usage of multiple promoters of the transactivator CIITA. EMBO J. 16, 2851–2860 (1997).
Shin, J.S. et al. Surface expression of MHC class II in dendritic cells is controlled by regulated ubiquitination. Nature 444, 115–118 (2006).
Busch, R., Doebele, R.C., Patil, N.S., Pashine, A. & Mellins, E.D. Accessory molecules for MHC class II peptide loading. Curr. Opin. Immunol. 12, 99–106 (2000).
Pierre, P. & Mellman, I. Developmental regulation of invariant chain proteolysis controls MHC class II trafficking in mouse dendritic cells. Cell 93, 1135–1145 (1998).
Smith, M.A. et al. Positive regulatory domain I (PRDM1) and IRF8/PU.1 counter-regulate MHC class II transactivator (CIITA) expression during dendritic cell maturation. J. Biol. Chem. 286, 7893–7904 (2011).
Rescigno, M. et al. Bacteria-induced neo-biosynthesis, stabilization, and surface expression of functional class I molecules in mouse dendritic cells. Proc. Natl. Acad. Sci. USA 95, 5229–5234 (1998).
ten Broeke, T., van Niel, G., Wauben, M.H., Wubbolts, R. & Stoorvogel, W. Endosomally stored MHC class II does not contribute to antigen presentation by dendritic cells at inflammatory conditions. Traffic 12, 1025–1036 (2011).
Basu, S., Mehreja, R., Thiberge, S., Chen, M.T. & Weiss, R. Spatiotemporal control of gene expression with pulse-generating networks. Proc. Natl. Acad. Sci. USA 101, 6355–6360 (2004).
Glasmacher, E. et al. A genomic regulatory element that directs assembly and function of immune-specific AP-1-IRF complexes. Science 338, 975–980 (2012).
Hogquist, K.A. et al. T cell receptor antagonist peptides induce positive selection. Cell 76, 17–27 (1994).
Barnden, M.J., Allison, J., Heath, W.R. & Carbone, F.R. Defective TCR expression in transgenic mice constructed using cDNA-based alpha- and beta-chain genes under the control of heterologous regulatory elements. Immunol. Cell Biol. 76, 34–40 (1998).
Battegay, M. et al. Quantification of lymphocytic choriomeningitis virus with an immunological focus assay in 24- or 96-well plates. J. Virol. Methods 33, 191–198 (1991).
Bettadapura, J., Menon, K.K., Moritz, S., Liu, J. & Bernard, C.C. Expression, purification, and encephalitogenicity of recombinant human myelin oligodendrocyte glycoprotein. J. Neurochem. 70, 1593–1599 (1998).
Lindstedt, M., Lundberg, K. & Borrebaeck, C.A. Gene family clustering identifies functionally associated subsets of human in vivo blood and tonsillar dendritic cells. J. Immunol. 175, 4839–4846 (2005).
Ritchie, M.E. et al. A comparison of background correction methods for two-colour microarrays. Bioinformatics 23, 2700–2707 (2007).
Bolstad, B.M., Irizarry, R.A., Astrand, M. & Speed, T.P. A comparison of normalization methods for high density oligonucleotide array data based on variance and bias. Bioinformatics 19, 185–193 (2003).
Smyth, G.K. Linear models and empirical bayes methods for assessing differential expression in microarray experiments. Statistical applications in genetics and molecular biology 3, Article3 (2004).
Acknowledgements
We thank members of the Singh and Mellman laboratories, as well as D. Holmes, for discussions and suggestions; and the animal husbandry technicians and flow cytometry, microarray, DNA sequencing, microscopy and oligonucleotide synthesis facilities at Genentech for assistance and reagents.
Author information
Authors and Affiliations
Contributions
A.A.K. and J.A.H. performed bioinformatics analyses; S.A. performed ChIP-Seq analysis; J.L. and J.D. assisted with animal studies; M.Z. and W.P.L. performed TNP-OVA airway-inflammation studies; S.P. and M.X. performed LCMV studies; C.J.S. assisted with ChIP-Seq and transduction studies; C.C. performed immunofluorescent microscopy; L.D. and I.M. assisted with design of antigen-presentation studies; B.V.L. and H.S. designed the study, analyzed data and prepared the manuscript. All authors discussed the results and commented on the manuscript.
Corresponding author
Ethics declarations
Competing interests
B.V.L., J.A.H., S.A., J.L., M.Z., W.P.L., S.P., M.X., J.D., C.J.S., C.C., L.D., I.M. and H.S. were employees of Genentech at the time of this work.
Integrated supplementary information
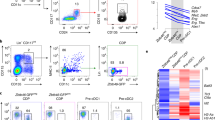
Supplementary Figure 1 Selective loss of CD11b+ DCs in Irf4fl/fl Cd11c-Cre mice.
(a) Splenic DC populations of wild-type mice were analyzed by flow cytometry for intracellular expression of IRF4 and IRF8 protein. Filled grey histograms are isotype controls. IRF4 and IRF8 expression was similarly analyzed for MHCIIhi DC populations in cutaneous lymph nodes (CLN) (d) and mesenteric lymph nodes (MLN) (g). (b) Left, DC-enriched cells from spleens of Cd11c-Cre and Irf4fl/fl Cd11c-Cre mice were analyzed by flow cytometry for expression of CD11c and MHCII. Numbers indicate percentage of cells within the CD11c+MHCIIint gate. Right, CD11c+MHCIIint DC subpopulations were distinguished by expression of CD11b and CD103. Numbers indicate percentage of cells within each gate. (c) Quantitative analysis of DCs described in (b). Data represent mean ± s.e.m. from 3-5 independent experiments, each with pooled splenic tissue from 5 animals. NS = not significant; * = P < 0.05, two-tailed t-test. (e) Left, DC-enriched cells from CLNs of Cd11c-Cre and Irf4fl/fl Cd11c-Cre mice were analyzed by flow cytometry for expression of CD11c and MHCII. Numbers indicate percentage of cells within the CD11c+MHCIIhi gate. Right, CD11c+MHCIIhi migratory DC subpopulations were distinguished by expression of CD11b and CD103. Numbers indicate percentage of cells within each gate. (f) Quantitative analysis of DCs described in (e). Data represent mean ± s.e.m. from at least 3 independent experiments, each with pooled CLN tissue from 5 animals. NS = not significant; * = P < 0.05, two-tailed t-test. (h, i) Migratory MHCIIhi DCs from MLN of Cd11c-Cre and Irf4fl/fl Cd11c-Cre mice were analyzed as in panels e and f. Data represent mean ± s.e.m. from at least 3 independent experiments, each with pooled CLN tissue from 5 animals. NS = not significant; * = P < 0.05, two-tailed t-test.
Supplementary Figure 2 IRF4-dependent CD11b+ DCs preferentially prime helper T cell responses to cell-associated antigen.
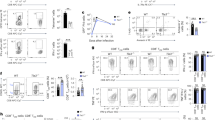
CellTrace violet-labeled OT-I and OT-II T cells were co-transferred into Cd11c-Cre and Irf4fl/fl Cd11c-Cre mice. Recipient mice were subsequently immunized by footpad injection with the indicated numbers of necrotic ovalbumin-expressing MC38 (MC38-OVA) tumor cells. Histograms show OT-I and OT-II proliferation measured by dye dilution, 3 days after antigen administration. For each dose, paired OT-II and OT-I data from the same mouse is displayed. Data are representative of 2 independent experiments, n = 2 mice per dose in each experiment.
Supplementary Figure 3 Selective impairment of TH-dependent immunity in Irf4fl/fl Cd11c-Cre mice.
(a, b) Cd11c-Cre and Irf4fl/fl Cd11c-Cre mice were immunized with TNP-OVA and serum antibody titers for IgG1 (a) and IgE (b) were determined by ELISA on the indicated days. Data represent antibody titers from 4 individual mice per group with horizontal bars indicating mean values. Data are representative of 2 independent experiments. (c, d) Cd11c-Cre and Irf4fl/fl Cd11c-Cre mice were sensitized with TNP-OVA. 35 days after sensitization mice were challenged by administration of nebulized TNP-OVA into the airways for 7 consecutive days. 24 h after the last antigen challenge lungs were removed. Lungs from naïve mice served as controls. Lung infiltrating CD4+ T cells (c) and neutrophils (d) were enumerated by flow cytometry. Data represent cell counts from individual mice (naïve n = 5, challenged n = 8) mean ± s.d.; one experimental replicate. (e, f) Cd11c-Cre and Irf4fl/fl Cd11c-Cre mice were infected with LCMV. Uninfected (UI) wild-type (WT) C56BL/6 mice served as controls. (e) 8 days after infection splenocytes were stimulated in vitro with LCMV gp33 peptide. CD8+ CTLs were analyzed by intracellular flow cytometry for expression of IFN-γ. Representative data from individual mice are shown. Numbers indicate percentage of gated CD8+IFN-γ+ splenocytes. (f) Viral titers in livers of infected mice were determined at day 8 after infection by pfu assay. Data depict titers determined for individual mice, mean indicated by horizontal bars; NS = not significant, one-tailed Mann-Whitney. Data are representative of 2 independent experiments.
Supplementary Figure 4 IRF4 as a regulatory determinant of enhanced pMHCII formation in murine and human DCs.
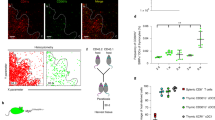
(a) Efficiency of pMHCII formation by MHCIIhiCD11b+CD103– (red) or MHCIIhiCD11b–CD103+ (blue) CLN migratory DC subsets was assessed after subcutaneous administration of the indicated doses of Eα antigen in wild-type (WT) C57BL/6 mice. DCs were enriched and then stained, 18 h after injection of Eα, with Y-Ae antibody and analyzed by flow cytometry. Data are from two independent experiments, n = 2 mice per dose, and represent geometric mean fluorescence intensity (gMFI) ± s.e.m.; * = P < 0.05, one-tailed t-test. (b) Differential gene expression analysis in paired mouse and human DC subsets. Mouse resident DCs are represented by splenic CD4+CD8– (CD4) and CD4–CD8+ (CD8) subsets. Mouse migratory DCs are represented by cutaneous lymph node MHCIIhiCD103–CD11b+ (CD11b) and MHCIIhiCD103+CD11b– (CD103) subsets. Human DCs isolated from blood are represented by BDCA1+ and BDCA3+ subsets. The murine data is from the ImmGen database17 whereas the human data is from E-TABM-34 (ref. 36). Expression of indicated genes is displayed as fold-increase (red) or decrease (blue) relative to its average expression in paired DC subsets.
Supplementary Figure 5 Flt3L-generated BMDCs express abundant IRF8 but not IRF4 and are not a suitable model for IRF4-dependent CD11b+ DCs.
BMDCs were differentiated from Cd11c-Cre (a) and Irf4fl/fl Cd11c-Cre (b) hematopoietic progenitors by culture in the presence of Flt3L. BMDCs were analyzed by flow cytometry for their expression of CD11c and MHCII. Numbers indicate percentages of cells with the CD11c+MHCII+ DC gate. CD11c+MHCII+ DC subpopulations were distinguished by their expression of CD11b. Numbers indicate percentage of cells within each gate. CD11b+ (red) and CD11b– (blue) DC populations were analyzed by flow cytometry for intracellular expression of IRF4 or IRF8 protein. Filled grey histograms are isotype controls. Data are representative of 2 independent experiments.
Supplementary Figure 6 IRF4 regulates the DC maturation program and MHCII antigen presentation.
(a-c) BMDCs were differentiated from Cd11c-Cre hematopoietic progenitors by culture in the presence of GM-CSF and IL-4. (a) CD11c+ DC populations were analyzed by flow cytometry for intracellular expression of IRF4 or IRF8 protein. Filled grey histograms are isotype controls. Data are representative of 3 independent experiments. (b) BMDCs derived from Cd11c-Cre or Irf4fl/fl Cd11c-Cre mice were left unstimulated (US, dashed histrograms) or stimulated overnight with LPS (solid histograms), then analyzed by flow cytometry for expression of the indicated markers (see also Fig. 4a). Isotype controls are shown in filled grey histograms. Data are representative of at least 3 independent experiments. (c) Cd11c-Cre and Irf4fl/fl Cd11c-Cre BMDCs were loaded with the indicated concentrations of ovalbumin (OVA) then stimulated with LPS. Antigen-loaded CD11c+ DCs were isolated by magnetic separation and used to stimulate OT-II T cells for 3 days. Histograms show OT-II proliferation as measured by dilution of CellTrace violet dye. Data are representative of at least 3 independent experiments.
Supplementary Figure 7 IRF4 directly targets key genes for MHCII antigen presentation.
(a-c) Wild-type C56BL/6 BMDCs differentiated with GM-CSF and IL-4 were stimulated for 6 h with LPS. IRF4, PU.1, BATF, or JunB-bound chromatin fragments were immunoprecipitated and subjected to high-throughput sequencing (ChIPseq). Sequence tracks displaying peaks for each of the above transcription factors are shown for Ctss (a), H2-Dmb2 (b), and Ciita (c) genes. Boxed peaks containing Ets-IRF (EICE) or AP-1-IRF (AICE) composite motifs are highlighted. Two independent ChIPseq replicates showed greater than 95% overlap in called IRF4 binding sites.
Supplementary Figure 8 IRF4 and IRF8 comparably induce aspects of DC maturation.
(a) Irf4fl/fl Cd11c-Cre hematopoietic progenitors were transduced with MSCV-IRES-huCD4 (MSCV) or its IRF4- or IRF8-expressing derivatives and then differentiated into DCs with GM-CSF and IL-4 (see Fig. 5a). Complemented BMDCs were analysed by flow cytometry for expression of IRF4, IRF8, CD80, and CD86. Histograms represent staining for MSCV control (black), MSCV-IRF4 (red), and MSCV-IRF8 (blue) complemented BMDCs. Data are representative of at least 3 independent experiments. (b) Complemented BMDCs were loaded with the indicated concentrations of ovalbumin (OVA) protein and then stimulated overnight with LPS. Transduced CD11c+ cells were purified by flow cytometry and used to stimulate labelled OT-I T cells for 3 days. T cell proliferation was analysed by dye dilution. Data are representative of 2 independent experiments.
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–8 (PDF 4469 kb)
Rights and permissions
About this article
Cite this article
Vander Lugt, B., Khan, A., Hackney, J. et al. Transcriptional programming of dendritic cells for enhanced MHC class II antigen presentation. Nat Immunol 15, 161–167 (2014). https://doi.org/10.1038/ni.2795
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ni.2795
This article is cited by
-
Signaling pathways involved in the biological functions of dendritic cells and their implications for disease treatment
Molecular Biomedicine (2023)
-
Early life exposure to house dust mite allergen prevents experimental allergic asthma requiring mitochondrial H2O2
Mucosal Immunology (2022)
-
Dendritic cell migration in inflammation and immunity
Cellular & Molecular Immunology (2021)
-
Dendritic cell biology and its role in tumor immunotherapy
Journal of Hematology & Oncology (2020)
-
U-DCS: characterization of the first permanent human dendritic sarcoma cell line
Scientific Reports (2020)