Abstract
The NLRP3 inflammasome functions as a crucial component of the innate immune system in recognizing viral infection, but the mechanism by which viruses activate this inflammasome remains unclear. Here we found that inhibition of the serine-threonine kinases RIP1 (RIPK1) or RIP3 (RIPK3) suppressed RNA virus–induced activation of the NLRP3 inflammasome. Infection with an RNA virus initiated assembly of the RIP1-RIP3 complex, which promoted activation of the GTPase DRP1 and its translocation to mitochondria to drive mitochondrial damage and activation of the NLRP3 inflammasome. Notably, the RIP1-RIP3 complex drove the NLRP3 inflammasome independently of MLKL, an essential downstream effector of RIP1-RIP3–dependent necrosis. Together our results reveal a specific role for the RIP1-RIP3-DRP1 pathway in RNA virus–induced activation of the NLRP3 inflammasome and establish a direct link between inflammation and cell-death signaling pathways.
Similar content being viewed by others
Main
The Nod-like receptor NLRP3, together with the adaptor ASC and caspase-1, can form a protein complex called the 'NLRP3 inflammasome', which is responsible for the maturation and secretion of proinflammatory cytokines, such as interleukin 1β (IL-1β) and IL-18 (refs. 1,2). The NLRP3 inflammasome is activated by a wide range of stress signals that derive not only from pathogens but also from environmental stress, metabolic dysregulation and tissue damage. So, in addition to its critical role in host defense against pathogens, the NLRP3 inflammasome is involved in the progression of various inflammatory human diseases, such as gout and type 2 diabetes1,2,3. Although evidence has shown that potassium efflux, mitochondria dysfunction and lysosomal disruption are involved in activation of the NLRP3 inflammasome4,5,6,7,8, the precise mechanisms and signaling pathway of this activation remain poorly understood.
As a crucial component of the innate immune system, the inflammasome serves an important role in host defense by recognizing viral infection and triggering responses from the innate immune system9,10,11. Some DNA viruses, such as vaccinia virus and mouse cytomegalovirus, can stimulate activation of the AIM2 inflammasome via direct binding between double-stranded DNA and AIM2 (ref. 12). The NLRP3 inflammasome can recognize both RNA viruses and some DNA viruses, including adenovirus, influenza virus, Sendai virus and vesicular stomatitis virus (VSV)9,13,14,15. Although published reports have suggested that NLRP3 detects viral infection through ion flux generated by the virus-encoded M2 ion channel or through the recognition of viral RNA10,14,16,17,18, the mechanism by which NLRP3 detects viral infection remains unclear, in contrast to the mechanisms known for the recognition of DNA viruses by AIM2.
The serine-threonine kinase RIP3 (RIPK3) is a member of the RIP family that functions as a key participant in programmed necrosis by forming a necrosome protein complex with RIP1 (refs. 19,20,21). In addition to their central role in necrosis, the necrosome components RIP1 and RIP3 have been proposed to be involved in the activation of inflammasomes22,23,24,25,26. When inhibitory factors of necrosomes, such as members of the IAP family or caspase-8, are inhibited, RIP1 and RIP3 can promote spontaneous activation of the inflammasome22,23,24. In addition, the activation of inflammasomes induced by infection with Yersinia pestis depends on RIP1 but not RIP3 (refs. 25,26), which suggests that the role of RIP1 in bacteria-induced activation of inflammasomes is independent of necrosomes. In mice with mutant tyrosine-phosphatase Ptpn6, RIP1 drives autoinflammation by promoting IL-1α production without the involvement of RIP3 or the inflamamsome27. Thus, although more and more evidence suggests a close connection between inflammation and necrosis, it is still unclear whether the RIP1-RIP3 complex can contribute to the activation of inflammasomes in physiological or pathological conditions. In addition, although the mechanism by which the RIP1-RIP3 complex induces necrosis is emerging, how this complex signals activation of the inflammasome and inflammation is unclear. In this study, we found that the RIP1-RIP3 complex was involved in RNA virus–induced activation of the NLRP3 inflammasome. Infection with an RNA virus initiated assembly of the RIP1-RIP3 complex, which activated the GTPase DRP1 and promoted mitochondrial damage, production of reactive oxygen species (ROS) and activation of the NLRP3 inflammasome.
Results
RIP3 promotes RNA virus–induced NLRP3 activation
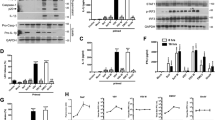
To assess the role of RIP3 in inflammasome activation, we treated bone marrow–derived macrophages (BMDMs) from RIP3-deficient (Ripk3−/−; called 'Rip3−/−' here) mice with classic NLRP3 agonists, including ATP, monosodium urate (MSU), aluminium sulfate (alum) and nigericin. In these conditions, IL-1β secretion was similar in Rip3−/− cells and Rip3+/+ cells (Fig. 1a). Similarly, RIP3 deficiency had no effect on activation of the AIM2 inflammasome by treatment with the synthetic double-stranded DNA poly(dA:dT) (Fig. 1a), which suggested that RIP3 was not involved in inflammasome activation induced by classic agonists of NLRP3 or AIM2. When we investigated whether RIP3 was involved in virus-induced activation of the inflammasome, we found that VSV-induced secretion of IL-1β was suppressed in BMDMs from both Nlrp3−/− mice and Rip3−/− mice compared with its secretion in wild-type cells (Fig. 1b). The secretion of IL-18 and cleavage of caspase-1 were also reduced in Nlrp3−/− and Rip3−/− BMDMs compared with that in wild-type cells (Fig. 1c,d), which suggested a role for RIP3 in promoting VSV-induced activation of the NLRP3 inflammasome. To exclude the possibility that the impaired IL-1β secretion in Rip3−/− cells might have been due to less cell death, we investigated whether RIP3 deficiency affected cell death induced by viral infection. Infection with VSV induced minimal death of both wild-type cells and Rip3−/− cells (Supplementary Fig. 1a). In addition, VSV-induced production of interferon-β was not affected by RIP3 deficiency (Supplementary Fig. 1b), which suggested a specific role for RIP3 in activation of the NLRP3 inflammasome.
(a) Secretion of IL-1β by LPS-primed Rip3+/+ or Rip3−/− BMDMs left unstimulated (US) or stimulated for 30 min with ATP or nigericin or for 4 h with MSU, alum or poly(dA:dT). ND, not detectable. (b,c) Secretion of IL-1β (b) and IL-18 (c) by LPS-primed wild-type (WT), Rip3−/− or Nlrp3−/− BMDMs infected for 6 h with various doses of VSV (multiplicity of infection (MOI), horizontal axis). (d) Immunoblot analysis of IL-1β and cleaved caspase-1 (p20) in culture supernatants (SN) of LPS-primed wild-type, Rip3−/− or Nlrp3−/− BMDMs infected for 6 h with various doses (above lanes) of VSV, and immunoblot analysis of the precursors of IL-1β (pro-IL-1β) and caspase-1 (pro-casp1) in lysates of those cells (Input). (e,f) Secretion of IL-1β by LPS-primed Nlrp3+/+ or Nlrp3−/− BMDMs (e) or Rip3+/+ or Rip3−/− BMDMs (f) left uninfected (UI) or infected for 6 h with VSV or adenovirus (AdV) or for 24 h with Sendai virus (SeV), influenza virus (Flu) or HSV. NS, not significant (P > 0.05; unpaired t-test (a) or two-way analysis of variance (ANOVA) (f)); *P < 0.05 and **P < 0.001 (two-way ANOVA). Data are from three independent experiments with biological duplicates in each (a–c,e,f; mean and s.e.m. of n = 6 (duplicates in three experiments)) or are representative of at least three independent experiments (d).
We further investigated whether RIP3 was involved in activation of the NLRP3 inflammasome induced by other viruses. Consistent with published reports10,14,15, RNA viruses, such as Sendai virus and influenza virus, stimulated IL-1β secretion in macrophages in an NLRP3-dependent manner (Fig. 1e). Similarly, IL-1β production induced by VSV, Sendai virus or influenza virus was significantly impaired in Rip3−/− macrophages compared with that in wild-type cells (Fig. 1f), which suggested a role for RIP3 in RNA virus–induced activation of the inflammasome. IL-1β secretion induced by DNA viruses such as adenovirus or herpes simplex virus (HSV) was also NLRP3 dependent (Fig. 1e). However, in contrast to the induction of its production by RNA viruses, IL-1β production induced by adenovirus or HSV was RIP3 independent (Fig. 1f).
We next investigated the role of RIP3 in activation of the NLRP3 inflammasome during in vivo infection with RNA viruses. Infection with influenza virus, VSV or HSV resulted in a higher concentration of IL-1β and IL-18 in the bronchoalveolar lavage fluid (BALF) of wild-type (Rip3+/+) mice than did mock infection (Fig. 2a,b). Infection with influenza virus or VSV induced a lower concentration of IL-1β and IL-18 in the BALF of Rip3−/− mice than in that of wild-type mice, while the concentration of IL-1β and IL-18 induced by infection with HSV was not significantly different in Rip3−/− mice versus wild-type mice (Fig. 2a,b); this suggested that RIP3 was required for RNA virus–induced activation of the NLRP3 inflammasome in vivo. However, the concentration of IL-6, an inflammasome-independent cytokine, was similar in Rip3−/− mice and wild-type mice in all infection conditions (Fig. 2c). Together these results indicated involvement of RIP3 in RNA virus–activated activation of the NLRP3 inflammasome.
Concentrations of IL-1β (a), IL-18 (b) and IL-6 (c) in BALF from Rip3+/+ or Rip3−/− mice (n = 11 per genotype) left uninfected or infected with influenza virus, VSV or HSV, assessed 3 d after infection. NS, not significant; *P < 0.001 (two-way ANOVA). Data are from three independent experiments (mean and s.e.m.).
RNA virus–induced NLRP3 activation depends on RIP1
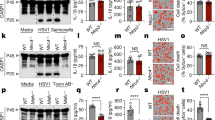
In programmed necrosis, activation of RIP3 is initiated by assembly of the necrosome with RIP1 (refs. 19,20). To address whether RIP1 was required for virus-induced activation of the NLRP3 inflammasome, we knocked down expression of RIP1 mRNA in THP-1 human macrophages with short hairpin RNA (shRNA) constructs. VSV-induced secretion of IL-1β and cleavage of caspase-1 were reduced in THP-1 cells in which RIP1 was silenced compared with that in cells in which it was not silenced (Fig. 3a,b). Similarly, knockdown of Rip1 mRNA by small interfering RNA (siRNA) in BMDMs inhibited VSV-induced IL-1β secretion but did not affect the priming effects of the Toll-like receptor 2 (TLR2) agonist Pam3CSK4 (Fig. 3c and Supplementary Fig. 1c,d). Furthermore, inhibition of RIP1's kinase activity with specific inhibitors of RIP1 (Nec-1 or Nec-1s)28 also suppressed VSV-induced IL-1β secretion, while the control inhibitor Nec-1i and the indoleamine 2,3-deoxygenase inhibitor 1-MT had no effect (Fig. 3d). These results indicated that RIP1 was important for RNA virus–induced activation of the NLRP3 inflammasome. Consistent with that, infection with VSV promoted binding of RIP1 to RIP3 in BMDMs (Fig. 3e), which suggested that viral infection initiated formation of the RIP1-RIP3 complex. These results indicated that formation of the RIP1-RIP3 complex promoted activation of the NLRP3 inflammasome during infection with an RNA virus.
(a) Immunoblot analysis of IL-1β and cleaved caspase-1 in supernatants of THP-1 cells stably expressing control shRNA with a scrambled sequence (Scr) or RIP1-specific shRNA (shRIP1) and infected for 6 h with various doses (above lanes) of VSV (top), and immunoblot analysis of pro-IL-1β, RIP1 and β-actin (loading control throughout) in lysates of those cells (bottom). (b) Secretion of IL-1β by THP-1 cells stably expressing shRNA and infected with VSV as in a. (c) Secretion of IL-1β by Pam3CSK4-primed BMDMs transfected with control siRNA with a scrambled sequence (Scr) or Rip1-specific siRNA (either of two constructs, siRip1 (1) or siRip1 (2)) and left untreated (UT) or stimulated for 4 h with MSU or infected for 6 h with VSV. (d) Secretion of IL-1β by LPS-primed BMDMs pretreated for 1 h with various inhibitors (below plot) and left uninfected (−) or infected (+) for 6 h with VSV. (e) Immunoblot analysis of RIP1 or RIP3 in immunoprecipitates (IP) or lysates (Input) of LPS-primed BMDMs infected with for 1 h with various doses of VSV (above lanes). (f,g) Secretion of IL-1β by LPS-primed Mlkl+/+ or Mlkl−/− BMDMs left untreated or infected with for 6 h VSV (f) or stimulated for 4 h with MSU or alum or for 30 min with ATP or nigericin (g). (h) Survival of wild-type, Rip3−/− or Mlkl−/− BMDMs treated for 24 h with the caspase inhibitor z-VAD-fmk (zVAD) alone (20 μM) or with z-VAD-fmk (20 μM) plus VSV (MOI, 3), assessed as the release of lactate dehydrogenase. NS, two-way ANOVA (c,d,g) or unpaired t-test (f); *P < 0.05, **P < 0.01 and ***P < 0.001 (two-way ANOVA (b,c,h) or unpaired t-test (d)). Data are representative of at least three independent experiments (a,e) or are from three independent experiments with biological duplicates in each (b–d,f–h; mean and s.e.m. of n = 6).
The RIP1-RIP3 complex induces necrosis through sequential activation of the effector MLKL and the phosphatase PGAM5 (refs. 29,30). However, secretion of IL-1β induced by VSV or classic agonists of NLRP3 was normal in MLKL-deficient (Mlkl−/−) BMDMs (Fig. 3f,g), although VSV-induced necrosis was blocked in Mlkl−/− BMDMs (Fig. 3h). Similarly, knockdown of PGAM5 mRNA in THP-1 cells had no effect on activation of the NLRP3 inflammasome induced by viral infection or other agonists (Supplementary Fig. 1e–g). These results indicated that MLKL and PGAM5 were not involved in virus-induced activation of the NLRP3 inflammasome downstream of the RIP1-RIP3 complex, although MLKL was essential for RNA virus–induced necrosis, which suggested that the RIP1-RIP3 complex induced cell death and inflammation via distinct downstream pathways.
The RIP1-RIP3 complex promotes mitochondrial damage
Mitochondrial dysfunction is involved in activation of the NLRP3 inflammasome induced by various agonists, including RNA viruses4,5,6,31,32,33. Because mitochondrial fission has been proposed to be downstream of the RIP1-RIP3 complex in promoting necrosis30 and because ROS produced by damaged mitochondria are important regulators of RNA virus–induced activation of the NLRP3 inflammasome34,35, we investigated whether RIP1 or RIP3 was involved in RNA virus–induced mitochondrial damage and ROS production. Similar to other NLRP3 agonists4, infection with VSV promoted mitochondrial fission, induction of ROS and the formation of mitochondrial aggregates in the perinuclear space of wild-type BMDMs (Fig. 4a,b and Supplementary Fig. 1h), and VSV-induced mitochondria fission, ROS production and aggregate formation were impaired in Rip3−/− BMDMs (Fig. 4a,b and Supplementary Fig. 1h). Inhibition of RIP1 activity with Nec-1s diminished the VSV-induced mitochondria fission, ROS production and aggregate formation in wild-type BMDMs (Fig. 4c,d and Supplementary Fig. 1i). In contrast, nigericin-induced mitochondrial ROS production and aggregate formation were not affected when RIP1 or RIP3 was inhibited (Fig. 4a–d and Supplementary Fig. 1h,i), which suggested a specific role for the RIP1-RIP3 complex in RNA virus–induced mitochondria damage. Furthermore, VSV-induced colocalization of ASC with mitochondria was inhibited in Rip3−/− BMDMs compared with their colocalization in wild-type cells (Fig. 4e and Supplementary Fig. 2). Together these results indicated that the RIP1-RIP3 complex was able to promote aberrant fission and damage of mitochondria to activate the NLRP3 inflammasome during infection with an RNA virus.
(a) Confocal microscopy of LPS-primed Rip3+/+ or Rip3−/− BMDMs (left margin) left untreated or infected for 2 h with VSV or stimulated for 30 min with nigericin (above images) and then stained with red fluorescent mitochondrial dye MitoTracker Red and with the DNA-binding dye DAPI (blue). Scale bar, 25 μm. (b) Quantification of cells (treated as in a) with mitochondrial aggregates in the perinuclear space ('mito aggregates'). (c) Confocal microscopy of LPS-primed BMDMs given no pretreatment (Mock) or pretreated for 1 h with Nec-1s (30 μM) (left margin), followed by treatment (above images) and staining as in a. Scale bar, 25 μm. (d) Quantification of cells treated as in c, assessed as in b. (e) Quantification of the colocalization of ASC and mitochondria ('ASC-mito') as assessed by confocal microscopy of LPS-primed Rip3+/+ or Rip3−/− BMDMs left uninfected or infected for 2 h with VSV and then stained with antibody to ASC, MitoTracker Red and DAPI. *P < 0.001 (two-way ANOVA). Data are representative of three experiments (a,c) or are from three independent experiments with biological duplicates in each (b,d,e; mean and s.e.m. of n = 6).
RIP1 and RIP3 induce translocation of DRP1 to mitochondria
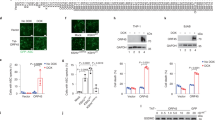
The GTPase DRP1 catalyzes the process of mitochondrial fission. Because infection with an RNA virus promoted aberrant mitochondrial fission, we investigated whether infection with VSV promoted activation of DRP1. Phosphorylation at Ser616 induces activation of DRP1 and its translocation to mitochondria, which then stimulates mitochondrial fission36. We observed that infection with VSV induced phosphorylation of DRP1 at Ser616 in wild-type BMDMs, while infection with adenovirus or treatment with other NLRP3 agonists did not (Fig. 5a); this suggested that infection with an RNA virus was able to activate DRP1. Moreover, VSV-induced phosphorylation of DRP1 was impaired in Rip3−/− BMDMs relative to such phosphorylation in wild-type BMDMs (Fig. 5b). Inhibition of RIP1 kinase activity by Nec-1 or Nec-1s inhibited the VSV-induced phosphorylation of DRP1 (Fig. 5c). In contrast, VSV-induced phosphorylation of DRP1 was similar in Mlkl−/− BMDMs and wild-type BMDMs (Fig. 5d), which suggested that the RNA virus–induced activation of DRP1 depended on RIP1 and RIP3 but did not depend on the necrosis effector MLKL. Furthermore, VSV-induced translocation of DRP1 to mitochondria in BMDMs was inhibited following deletion of RIP3 or inhibition of RIP1 by Nec-1s (Supplementary Fig. 3 and Supplementary Fig. 4) but was not inhibited by deletion of MLKL (Supplementary Fig. 5). This suggested that the RIP1-RIP3 complex was able to activate DRP1 and promote its translocation to mitochondria during infection with an RNA virus.
(a) Immunoblot analysis of phosphorylated (p-) and total DRP1 in LPS-primed wild-type BMDMs left untreated (Mock) or stimulated for various times (above lanes) with MSU, nigericin or ATP or infected with various times (above lanes) with adenovirus or VSV. (b) Immunoblot analysis of phosphorylated and total DRP1 in LPS-primed Rip3+/+ or Rip3−/− BMDMs infected for various times (above lanes) with VSV. (c) Immunoblot analysis of phosphorylated and total DRP1 in LPS-primed BMDMs pretreated for 1 h with various inhibitors (above blots) and then left uninfected (−) or infected (+) for 30 min with VSV. (d) Immunoblot analysis of phosphorylated and total DRP1 in LPS-primed Mlk+/+ or Mlkl−/− BMDMs infected for various times (above lanes) with VSV. (e) Immunoblot analysis of the interaction of RIP1 or RIP3 with DRP1 in immunoprecipitates or lysates of HEK293T cells transfected to express enhanced green fluorescent protein (eGFP)-tagged DRP1 plus either empty vector (EV) or vector expressing Flag-tagged RIP1 or RIP3 (above lanes). (f) In vitro phosphorylation assay of DRP1 incubated with purified RIP1 or RIP3 from lysates of HEK293T cells transfected as in e, analyzed by immunoblot with antibody to DRP1 phosphorylated at Ser616. (g) Immunoblot analysis of DRP1, RIP1 and RIP3 in immunoprecipitates or lysates of LPS-primed Rip3+/+ or Rip3−/− BMDMs left uninfected (−) or infected (+) for 1 h with VSV. Data are representative of three independent experiments.
Next we investigated how RIP1 and RIP3 activated DRP1. When overexpressed in HEK293T human embryonic kidney cells, RIP1 interacted with DRP1, while overexpressed RIP3 did not (Fig. 5e). In an in vitro phosphorylation assay, RIP1 phosphorylated DRP1 at Ser616 (Fig. 5f), which suggested that RIP1 was able to interact with and phosphorylate DRP1 to promote its activation. Furthermore, infection with VSV promoted binding of RIP1 to RIP3 and DRP1 in BMDMs (Fig. 5g), which suggested that infection with VSV induced the formation of a RIP1-RIP3-DRP1 complex. The VSV-induced interaction between RIP1 and DRP1 was blocked in Rip3−/− BMDMs (Fig. 5g), which suggested that RIP3 regulated the interaction between RIP1 and DRP1 during infection with VSV. Together our results indicated that the RIP1-RIP3 complex was able to promote activation of DRP1 via interaction between RIP1 and DRP1 during infection with an RNA virus.
DRP1 is required for RNA virus–induced NLRP3 activation
Because the RIP1-RIP3 complex activated DRP1 and promoted its translocation to mitochondria, we next investigated the role of DRP1 in RNA virus–induced activation of the NLRP3 inflammasome. The VSV-induced formation of mitochondrial aggregates, mitochondrial fission and ROS production were impaired in BMDMs in which Drp1 expression was silenced by siRNA compared with those effects in BMDMs in which it was not silenced (Fig. 6a,b and Supplementary Fig. 6a,b). Consistent with that impaired mitochondrial damage and ROS production, VSV-induced production of IL-1β was also impaired in BMDMs in which Drp1 mRNA was silenced with siRNA compared with its production in BMDMs in which Drp1 mRNA was not silenced (Fig. 6c), while VSV-induced production of tumor-necrosis factor was not affected (Fig. 6d). Similar to the effect of RIP3 deficiency, inhibition of Drp1 expression with siRNA did not affect the DNA virus–induced production of IL-1β (Fig. 6c). In addition, knockdown of Drp1 mRNA had no effect on MSU-, ATP- or nigericin-induced activation of the NLRP3 inflammasome (Supplementary Fig. 6c), which suggested a specific role for DRP1 in RNA virus–induced activation of the inflammasome. The mitochondrial fission protein Fis1 is an important component of the mitochondrial outer membrane that can recruit DRP1 to mitochondria and promote mitochondrial fission37. Consistent with the effect of silencing Drp1, we found that silencing of Fis1 with siRNA in BMDMs inhibited VSV-induced mitochondrial damage and IL-1β secretion but had no effect on MSU- or nigericin-induced secretion of IL-1β (Fig. 6e,f and Supplementary Fig. 6d–f).
(a) Confocal microscopy of BMDMs transfected with control siRNA or Drp1-specific siRNA (left margin) and left untreated or infected for 2 h with VSV or stimulated for 30 min with nigericin (above images), followed by staining with MitoTracker Red and DAPI. Scale bar, 25 μm. (b) Quantification of cells treated as in a (assessed as in Fig. 4b). (c,d) Secretion of IL-1β (c) or tumor-necrosis factor (TNF) (d) by LPS-primed BMDMs transfected with siRNA as in a and then left uninfected or infected for 6 h with VSV or adenovirus. (e,f) Secretion of IL-1β by LPS-primed BMDMs transfected with control siRNA or Fis1-specific siRNA and left untreated or infected for 6 h with VSV (e) or stimulated for 4 h with MSU or for 30 min with nigericin (f). (g,h) Survival of LPS-primed BMDMs transfected with siRNA as in a (g) or e,f (h) and then stimulated for 24 h with z-VAD-fmk with or without VSV (as in Fig. 3h), assessed as release of lactate dehydrogenase. NS, two-way ANOVA; *P < 0.001 (two-way ANOVA). Data are representative of three independent experiments (a) or are from three independent experiments with biological duplicates in each (b–h; mean and s.e.m. of n = 6).
Dissipation of the membrane potential by CCCP, a chemical inhibitor of oxidative phosphorylation, can induce mitochondrial fission via DRP1 (ref. 38), so to further study the specificity of the RIP1-RIP3-DRP1 signaling pathway in RNA virus–induced activation of NLRP3, we investigatd the role of RIP3 in CCCP-induced mitochondria fission. CCCP did not induce phosphorylation of DRP1 at Ser616, and although CCCP induced IL-1β production and translocation of DRP1 to mitochondria in wild-type BMDMs, this was not affected by RIP3 deficiency (Supplementary Fig. 7a–c). However, CCCP-induced secretion of IL-1β was impaired when Drp1 expression was inhibited by siRNA in BMDMs (Supplementary Fig. 7d), consistent with a role for DRP1 in CCCP-induced mitochondrial fission (Supplementary Fig. 7e,f); this suggested that CCCP activated the NLRP3 inflammasome in a DRP1-dependent but RIP3-independent manner. Together these results indicated that DRP1 acted downstream of the RIP1-RIP3 complex and promoted RNA virus–induced activation of the NLRP3 inflammasome.
Next we found that knockdown of Drp1 mRNA in BMDMs did not suppress VSV-induced necrosis (Fig. 6g). Similarly, knockdown of Fis1 mRNA in BMDMs had no effect on VSV-induced necrosis (Fig. 6h). Together these results indicated that DRP1 acted downstream of the RIP1-RIP3 complex to promote mitochondrial damage and activation of the NLRP3 inflammasome, while MLKL acted downstream of RIP1 and RIP3 to induce necrosis; this suggested that the RIP1-RIP3 complex used different pathways to signal inflammation or necrosis during infection with an RNA virus.
Double-stranded RNA activates NLRP3 via RIP1 and RIP3
Because cytosolic RNA has been proposed to be the trigger of the NLRP3 inflammasome during infection with an RNA virus14,16,18, we investigated whether this process depends on RIP1 or RIP3. Similar to infection with an RNA virus, transfection of poly(I:C), a synthetic analog of double-stranded RNA (dsRNA), induced less secretion of IL-1β or IL-18 and cleavage of caspase-1 in Rip3−/− BMDMs than in wild-type BMDMs (Fig. 7a–c). In addition, knockdown of RIP1 mRNA in THP-1 cells inhibited the production of IL-1β induced by transfection of poly(I:C) compared with such production in THP-1 cells in which RIP1 mRNA was not silenced (Fig. 7d); this suggested that cytosolic RNA–induced activation of the NLRP3 inflammasome depended on the RIP1-RIP3 complex. Moreover, secretion of IL-1β or IL-18 induced by transfection of poly(I:C) into BMDMs was suppressed by siRNA-mediated knockdown of Drp1 or Fis1 mRNA compared with such secretion in BMDMs in which Drp1 or Fis1 mRNA was not silenced (Fig. 7e–h).These results indicated that the RIP1-RIP3-DRP1 pathway was important for cytosolic RNA–induced activation of the inflammasome.
(a,b) Secretion of IL-1β (a) and IL-18 (b) by LPS-primed wild-type, Rip3−/− or Nlrp3−/− BMDMs left unstimulated or stimulated for 6 h with poly(I:C). (c) Immunoblot analysis of IL-1β and cleaved caspase-1 in culture supernatants of LPS-primed wild-type, Rip3−/− or Nlrp3−/− BMDMs stimulated for 6 h with poly(I:C) (top), and immunoblot analysis of the precursors of IL-1β and caspase-1 in lysates of those cells (bottom). (d) Secretion of IL-1β by THP-1 cells stably expressing control shRNA or RIP1-specific shRNA and left unstimulated or stimulated for 6 h with poly(I:C). (e–h) Secretion of IL-1β (e,g) or IL-18 (f,h) by LPS-primed BMDMs transfected with control or Drp1-specific siRNA (e,f) or control or Fis1-specific siRNA (g,h) and then left unstimulated or stimulated for 6 h with poly(I:C). *P < 0.01 and **P < 0.001 (two-way ANOVA (a,b,e–h) or unpaired t-test (d)). Data are from three independent experiments with biological duplicates in each (a,b,d–h; mean and s.e.m. of n = 6) or are representative of three independent experiments (c).
RNA virus activates NLRP3 independently of RNA sensors
We next sought to determine what activates the NLRP3 inflammasome upstream of RIP1 and RIP3 during infection with an RNA virus and analyzed the role of known sensors that detect RNA in this process. Two cytosolic sensors of viral RNA, RIG-I and Mda5, induce antiviral innate immunity, but their role in virus-induced activation of inflammasomes is controversial15,39. IL-1β production induced by infection with VSV or cytosolic treatment with poly(I:C) was similar in RIG-I-deficient (Ddx58−/−; called 'Rig-I−/−' here) BMDMs and Mda5-deficient (Ifih1−/−; called 'Mda5−/−' here) BMDMs primed with lipopolysaccharide (LPS) compared with that in their wild-type counterparts (Supplementary Fig. 8a,b), although VSV-induced production of interferon-β was blocked in Rig-I−/− BMDMs (Supplementary Fig. 8c). DHX33 is a member of DExD/H-box (Asp–Glu–x–Asp or His, where 'x' is any amino acid) family of helicases and has been proposed to sense cytosolic dsRNA and activate the NLRP3 inflammasome during viral infection40. Similar to results in published reports in which knockdown of DHX33 mRNA in THP-1 cells resulted in only slightly decreased production of IL-1β induced by respiratory syncytial virus40, we found that shRNA-mediated knockdown of DHX33 mRNA in THP-1 cells had only a slight effect on the VSV-induced production of IL-1β, while silencing of RIP1 mRNA in these cells almost completely inhibited the VSV-induced production of IL-1β (Supplementary Fig. 8d,e); this suggested that DHX33 was not a major sensor of viral RNA–induced activation of NLRP3 during viral infection. We also assessed the role of TLR3, which senses extracellular dsRNA and activates innate immunity41, in RNA virus–induced activation of the NLRP3 inflammasome. Consistent with published reports16, VSV-induced production of IL-1β was not affected by deficiency in TLR3 (Supplementary Fig. 8f,g). Together these results suggested that RIG-I, Mda5, DHX33 and TLR3 were not the main sensors of RNA involved in RNA virus–induced activation of the NLRP3 inflammasome.
Discussion
The NLRP3 inflammasome can sense infection with either RNA viruses or DNA viruses to activate innate immune responses and has been proposed as a crucial component of host defense in viral infection9,10,11, but the mechanisms by which NLRP3 detects viral infection are still unclear. Here we demonstrated a role for the RIP1-RIP3 complex in RNA virus–induced activation of the NLRP3 inflammasome. Inactivation of RIP1 or RIP3 severely impaired such activation, including cleavage of caspase-1 and secretion of IL-1β and IL-18, but had no effect on DNA virus–induced activation of the NLRP3 inflammasome. We also identified DRP1 as the effector downstream of RIP1-RIP3 that promoted NLRP3 activation. Our results thus indicate that the RIP1-RIP3 complex is an important participant in innate immune and inflammatory signaling pathways during viral infection beyond those previously identified for necrosis.
Although increasing evidence has suggested that mitochondria serve as a platform for activation of the NLRP3 inflammasome by producing mitochondrial ROS, lipids or DNA in response to damage or 'stress'4,5,6,32,33, the signaling pathways upstream of mitochondrial damage have remained unclear. Our results have demonstrated a role for the RIP1-RIP3 complex in RNA virus–induced mitochondrial damage, ROS production and activation of the NLRP3 inflamamsome. Our results are consistent with those of published reports showing that RIP3 signaling is able to promote mitochondrial production of ROS21,23. In addition, RIP3-dependent activation of the inflammasome in macrophages in which IAP proteins are inhibited can be inhibited by ROS inhibitors22. Such results suggest that RIP1 and RIP3 function as important signaling molecules upstream of mitochondrial damage to promote activation of the NLRP3 inflammasome, at least during infection with an RNA virus or when IAP proteins are inhibited. However, the signaling pathways upstream of mitochondrial damage that are involved in activation of the NLRP3 inflammasome induced by other agonists need to be further identified.
MLKL and PGAM5 have been proposed to be the downstream effectors of the RIP1-RIP3 complex in necrosis29,30. Here, we found that MLKL and PGAM5 were not involved in RNA virus–induced activation of the NLRP3 inflammasome, although RNA virus–induced necrosis was indeed blocked in Mlkl−/− macrophages. However, MLKL and PGAM5 were required for spontaneous activation of the NLRP3 inflammasome in caspase-8-deficient dendritic cells because inhibition of the expression of MLKL or PGAM5 by siRNA reduced LPS-induced secretion of IL-1β in caspase-8-deficient dendritic cells22. Such results suggest that RIP1 and RIP3 might promote activation of the NLRP3 inflammasome via different signaling pathways in different 'stressed' conditions. The different roles and mechanisms of PGAM5 or MLKL in activation of the NLRP3 inflammasome needs to be further investigated in the future.
Our results have demonstrated that DRP1 acted downstream of RIP1 and RIP3 to activate the NLRP3 inflammasome by promoting mitochondrial fission during infection with an RNA virus. However, the roles of DRP1 and mitochondrial fission in inflammasome activation are complicated. Although nigericin did not induce phosphorylation of DRP1 at Ser616, it still induced the translocation of DRP1 to mitochondria, which suggests the existence of another mechanism for the activation of DRP1. Moreover, nigericin-induced mitochondrial damage, ROS production and inflammasome activation were normal when Drp1 was silenced, which indicates the existence of other pathways that regulate mitochondrial fission and mitochondrial damage during activation of the NLRP3 inflammasome induced by classic agonists. Indeed, DRP1-independent mitochondrial fission has been described38,42.
DRP1 has been proposed to be downstream of PGAM5 in triggering necrosis30. After the induction of necrosis by tumor-necrosis factor, PGAM5 might recruit DRP1 and activate its GTPase activity by dephosphorylating Ser637 of DRP1 to promote mitochondrial fission and necrosis30. However, we found that DRP1 was not involved in RNA virus–induced necrosis, consistent with a report showing that RIP3-dependent necrosis requires MLKL but not DRP1 (ref. 43). These discrepancies might be explained by differences in the stimuli or cell types used in these studies.
Although our results suggested a role for the RIP1-RIP3 complex in inflammasome activation induced by RNA viruses or dsRNA, the host sensor that detects viral RNA to trigger inflammasome activation remains unknown. RIG-I is a sensor of the innate immune system that detects cytosolic viral RNA, but its role in virus-induced inflammasome activation is controversial15,39. Published reports have suggested that RNA virus–induced production of IL-1β in unprimed bone marrow–derived dendritic cells is dependent on RIG-I but independent of NLRP3 (ref. 39). In contrast, another study has reported that such IL-1β production in LPS-primed BMDMs is dependent on NLRP3 but independent of RIG-I (ref. 15). Our results are consistent with the results of the latter study. These divergent results could be explained by the use of different stimulation protocols because it is possible that RIG-I might contribute to induction of the expression of NLRP3 or pro-IL-1β by viral infection in unprimed cells. In addition, our results indicated minimal roles for the RNA sensors DHX33 and TLR3 in RNA virus–induced activation of inflammasomes, which suggests that an undefined sensor of RNA might act upstream of the RIP1-RIP3 complex to promote activation of the NLRP3 inflammasome.
Collectively, our results have shown a role for the RIP1-RIP3 complex in RNA virus–induced activation of the NLRP3 inflammasome. This not only improves understanding of the mechanisms by which the NLRP3 inflammasome is activated but also establishes a direct link between inflammation and necrosis signaling pathways. Our identification of the RIP1-RIP3 complex as a crucial regulator in virus-induced inflammation might provide a potential new target for the treatment of inflammatory diseases associated with viral infection.
Methods
Mice.
Nlrp3−/−, Rip3−/−, Rig-I−/−, Mda5−/−, Mlkl−/− and Tlr3−/− mice have been described44,45,46,47,48,49. All mice were on a C57BL/6 background except that Rig-I−/− mice were on the BALB/c background. The sex-matched littermates of the mutant mice were used as controls. All animal experiments were approved by the Ethics Committee of University of Science and Technology of China.
Reagents.
Nigericin, CCCP (carbonyl cyanide m-chlorophenyl hydrazone), MSU, ATP, PMA (phorbol 12-myristate 13-acetate), z-VAD-fmk (benzyloxycarbonyl–Val-Ala-Asp–fluoromethylketone), poly(dA:dT) and 1-MT (1-methyl-[D]-tryptophan) were from Sigma. Nec-1s was from Biovision. Nec-1i was from Millipore. Nec-1 was from Cayman Chemical. Imject-Alum was from Pierce Biochemicals. Poly(I:C), Pam3CSK4 (tripalmitoyl cysteinyl seryl tetralysine lipopeptide) and ultrapure LPS were from Invivogen. MitoTracker and MitoSOX (fluorogenic dye specifically targeted to mitochondria in live cells) were from Invitrogen. The LDH (lactate dehydrogenase) cytotoxicity assay kit was from Beyotime. Protein G agarose was from Millipore. Antibody to human pro-IL-1β (60136-1-Ig) and antibody to Fis1 (anti-Fis1; 10956-1-AP) were from Proteintech. Antibody to mouse IL-1β (AF-401-NA) was from R&D Systems. Antibodies to mouse caspase-1 (p20) (AG-20B-0042) and anti-NLRP3 (AG-20B-0014) were from Adipogen. Anti-β-actin (P30002) was from Abmart. Anti-PGAM5 (186-200), anti-VSV (V4888) and anti-Flag (F2555) were from Sigma. Anti-RIP1 (3493), anti-DRP1 (8570), antibody to DRP1 phosphorylated at Ser616 (4494) and anti-human caspase-1 (2225) were from Cell Signaling. Anti-RIP3 (2283) was from ProSci. Anti-DHX33 (sc-137424) and anti-ASC (sc-22514-R) were from Santa Cruz. Antibody to human cleaved IL-1β (A5208206) was from Sangon Biotech.
Viruses.
VSV (Indiana strain) was propagated and amplified by infection of a monolayer of BHK-21 haby hamster kidney cells. HSV type 1 (KOS strain) was propagated on Vero African green monkey kidney cells. Influenza virus (strain A/PR/8/34) and Sendai virus were grown for 2 d at 37 °C in the allantoic cavities of 10-day-old fertile chicken eggs. Viruses in culture supernatants were quantified by standard plaque assay. Adenovirus was from Beijing Five Plus Molecular Medicine Institute. These viruses were stored at −80 °C.
Viral infection in vivo.
Anesthetized mice were challenged by intranasal administration of 3 × 103 plaque-forming units of influenza virus, 1 × 105 plaque-forming units of VSV or 1 × 105 plaque-forming units of HSV, each in 50 μl of PBS. At 3 d after infection, BALF was collected and was centrifuged for the removal of cells, and cytokines in the supernatant were quantified.
Generation of THP-1 cells expressing shRNA.
shRNA targeting mRNA of RIP1, PGAM5 and DHX33 was from Sigma. The protocol for generating THP-1 cells stably expressing shRNA has been published50.
siRNA-mediated gene silencing in BMDMs.
BMDMs were plated in 12-well plates (at a density of 4 × 105cells per well) and then were transfected with 50 nM siRNA through the use of Lipofectamine RNAiMAX according to the manufacturer's guidelines (Invitrogen). siRNA (sequences, Supplementary Table 1) was chemically synthesized by GenePharma, and the negative control siRNA was also from GenePharma.
Cell preparation and stimulation.
Human THP-1 cells were grown in RPMI-1640 medium supplemented with 10% FBS and 50 μM 2-mercaptoethanol. THP-1 cells from American Type Culture Collection were routinely tested for contamination by mycoplasma. THP-1 cells were differentiated for 3 h with 100 nM PMA and then were incubated overnight. BMDMs were derived from tibia and femoral bone marrow cells as described50 and were cultured in DMEM complemented with 10% FBS, 1 mM sodium pyruvate and 2 mM L-glutamine in the presence of culture supernatants of L929 mouse fibroblasts.
For induction of inflammasome activation, 5 × 105 macrophages were plated overnight in 12-well plates and the medium was changed to Opti-MEM (1% FBS) the following morning, then the cells were primed for 4 h with ultrapure LPS (100 ng/ml) or Pam3CSK4 (400 ng/ml). After that, the cells were stimulated for 4 h with MSU (150 μg/ml) or Alum (300 μg/ml) or for 30 min with ATP (2.5 mM) or nigericin (10 μM). Cells were transfected with poly(I:C) (1 μg/ml) or poly(dA:dT) (0.5 μg/ml) through the use of Lipofectamine according to the manufacturer's protocol (Invitrogen). For viral infection, primed BMDMs or differentiated THP-1 cells were infected for 6 h with VSV or adenovirus or for 24 h with Sendai virus, influenza virus or HSV type 1. If not indicated otherwise, cells were infected VSV, HSV type 1, influenza virus or Sendai virus at an MOI of 5 or with adenovirus at a dose of 1 × 104 particles/cell. Cell extracts and precipitated supernatants were analyzed by immunoblot.
Enzyme-linked immunosorbent assay.
Mouse IL-6 or IL-1β (R&D Systems), human IL-1β (BD Biosciences) or mouse IL-18 (eBioscience) in supernatants of cell culture or BALF were quantified by enzyme-linked immunosorbent assay according to the manufacturer's instructions.
Real time PCR.
BMDMs were dissolved in TRIzol reagent (Invitrogen). cDNA was synthesized from extracted total RNA with an M-MLV Reverse Transcriptase kit according to the manufacturer's protocol (Invitrogen). SYBR Green Premix (Takara) was used for quantitative PCR with a StepOne Real Time PCR System (Applied Biosystems). The gene encoding glyceraldehyde phosphate dehydrogenase was used as an internal control. The sequences of primers for quantitative PCR were as follows: interferon-β, 5′-CCAACAAGTGTCTCCTCCAAATT-3′ (forward) and 5′-GTAGGAATCCAAGCAAGTTGTAGCT-3′ (reverse); GAPDH, 5′-GGTGAAGGTCGGTGTGAACG-3′ (forward) and 5′-CTCGCTCCTGGAAGATGGTG-3′ (reverse).
Transfection and immunoprecipitation.
Constructs were transfected into HEK293T cells through the use of polyethylenimine. After 18 h, cells were collected and resuspended in lysis buffer (50 mM Tris, pH 7.8, 50 mM NaCl, 1% (vol/vol) Nonidet-P40, 5 mM EDTA and 10% (vol/vol) glycerol). Extracts were immunoprecipitated with anti-Flag (identified above) and beads and then were assessed by immunoblot analysis. LPS-primed BMDMs (1 × 107) were treated for 1 h with VSV. BMDMs were then resuspended in lysis buffer and proteins were immunoprecipitated from extracts with anti-RIP1 (identified above).
In vitro phosphorylation of DRP1.
Flag-tagged RIP1 or RIP3 was immunoprecipitated (with M2 (anti-Flag) beads; Sigma) from HEK293T cells transiently expressing these proteins. eGFP-DRP1 was immunoprecipitated from eGFP-DRP1–expressing HEK-293T cells with anti-eGFP (M20004; Abmart) and protein G agarose beads. After incubation for 4 h at 4 °C, the beads were collected and washed twice with ice-cold lysis buffer (described above) and once with ice-cold kinase buffer (50 mM Tris·HCl, pH 7.5, 5 mM MgCl2, 0.1 mM sodium orthovanadate, 0.1 mM sodium pyrophosphate, 1 mM NaF and 1 mM PMSF). Phosphorylation of DRP1 proceeded for 30 min at 37 °C in kinase buffer (final volume, 30 ml), ATP and immunoaffinity-purified RIP1 or RIP3. After separation of samples by SDS-PAGE, protein phosphorylation was detected with antibody to DRP1 phosphorylated at Ser616 (identified above).
Confocal microscopy.
BMDMs were plated on coverslips overnight and then were used for stimulation or staining with MitoTracker Red (50 nM) or MitoSOX (5 μM). After being washed three times with Tween-20 in PBS (PBST), the cells were fixed for 15 min with 4% PFA in PBS and then were washed three times with PBST. After permeabilization with Triton X-100 and blockade of nonspecific binding with 10% goat serum in PBS, cells were incubated overnight at 4 °C d with primary antibodies (identified above) in 10% goat serum. After being washed with PBST, cells were incubated for 60 min with Alexa Fluor 488–conjugated goat antibody to rabbit immunoglobulin G (A11008; Invitrogen) in 10% goat serum in PBS and were rinsed in PBST. A Zeiss LSM700 was used for confocal microscopy. For quantification of mitochondrial aggregates (>3 μm in diameter) in the perinuclear region or colocalization of ASC with mitochondria, scanning fields were randomly selected and at least 100 cells were counted in each slide.
Statistical analyses.
Statistical analyses were made with an unpaired t-test for two groups or two-way ANOVA (GraphPad Software) for multiple groups with all data points showing a normal distribution. Researchers were not blinded to the genotype of the mice or cells. No exclusion of data points was used. Sample sizes were selected on the basis of preliminary results to ensure an adequate power. P values of <0.05 were considered significant.
References
Chen, G., Shaw, M.H., Kim, Y.G. & Nunez, G. NOD-like receptors: role in innate immunity and inflammatory disease. Annu. Rev. Pathol. 4, 365–398 (2009).
Martinon, F., Mayor, A. & Tschopp, J. The inflammasomes: guardians of the body. Annu. Rev. Immunol. 27, 229–265 (2009).
Davis, B.K., Wen, H. & Ting, J.P. The inflammasome NLRs in immunity, inflammation, and associated diseases. Annu. Rev. Immunol. 29, 707–735 (2011).
Zhou, R., Yazdi, A.S., Menu, P. & Tschopp, J. A role for mitochondria in NLRP3 inflammasome activation. Nature 469, 221–225 (2011).
Iyer, S.S. et al. Mitochondrial cardiolipin is required for Nlrp3 inflammasome activation. Immunity 39, 311–323 (2013).
Misawa, T. et al. Microtubule-driven spatial arrangement of mitochondria promotes activation of the NLRP3 inflammasome. Nat. Immunol. 14, 454–460 (2013).
Hornung, V. et al. Silica crystals and aluminum salts activate the NALP3 inflammasome through phagosomal destabilization. Nat. Immunol. 9, 847–856 (2008).
Muñoz-Planillo, R. et al. K+ efflux is the common trigger of NLRP3 inflammasome activation by bacterial toxins and particulate matter. Immunity 38, 1142–1153 (2013).
Muruve, D.A. et al. The inflammasome recognizes cytosolic microbial and host DNA and triggers an innate immune response. Nature 452, 103–107 (2008).
Thomas, P.G. et al. The intracellular sensor NLRP3 mediates key innate and healing responses to influenza A virus via the regulation of caspase-1. Immunity 30, 566–575 (2009).
Kanneganti, T.D. Central roles of NLRs and inflammasomes in viral infection. Nat. Rev. Immunol. 10, 688–698 (2010).
Hornung, V. et al. AIM2 recognizes cytosolic dsDNA and forms a caspase-1-activating inflammasome with ASC. Nature 458, 514–518 (2009).
Ichinohe, T., Lee, H.K., Ogura, Y., Flavell, R. & Iwasaki, A. Inflammasome recognition of influenza virus is essential for adaptive immune responses. J. Exp. Med. 206, 79–87 (2009).
Allen, I.C. et al. The NLRP3 inflammasome mediates in vivo innate immunity to influenza A virus through recognition of viral RNA. Immunity 30, 556–565 (2009).
Rajan, J.V., Rodriguez, D., Miao, E.A. & Aderem, A. The NLRP3 inflammasome detects encephalomyocarditis virus and vesicular stomatitis virus infection. J. Virol. 85, 4167–4172 (2011).
Kanneganti, T.D. et al. Critical role for cryopyrin/Nalp3 in activation of caspase-1 in response to viral infection and double-stranded RNA. J. Biol. Chem. 281, 36560–36568 (2006).
Ichinohe, T., Pang, I.K. & Iwasaki, A. Influenza virus activates inflammasomes via its intracellular M2 ion channel. Nat. Immunol. 11, 404–410 (2010).
Rajan, J.V., Warren, S.E., Miao, E.A. & Aderem, A. Activation of the NLRP3 inflammasome by intracellular poly I:C. FEBS Lett. 584, 4627–4632 (2010).
Zhang, D.W. et al. RIP3, an energy metabolism regulator that switches TNF-induced cell death from apoptosis to necrosis. Science 325, 332–336 (2009).
He, S. et al. Receptor interacting protein kinase-3 determines cellular necrotic response to TNF-alpha. Cell 137, 1100–1111 (2009).
Cho, Y.S. et al. Phosphorylation-driven assembly of the RIP1–RIP3 complex regulates programmed necrosis and virus-induced inflammation. Cell 137, 1112–1123 (2009).
Kang, T.B., Yang, S.H., Toth, B., Kovalenko, A. & Wallach, D. Caspase-8 blocks kinase RIPK3-mediated activation of the NLRP3 inflammasome. Immunity 38, 27–40 (2013).
Vince, J.E. et al. Inhibitor of apoptosis proteins limit RIP3 kinase-dependent interleukin-1 activation. Immunity 36, 215–227 (2012).
Yabal, M. et al. XIAP Restricts TNF- and RIP3-dependent cell death and inflammasome activation. Cell Rep. 7, 1796–1808 (2014).
Philip, N.H. et al. Caspase-8 mediates caspase-1 processing and innate immune defense in response to bacterial blockade of NF-κB and MAPK signaling. Proc. Natl. Acad. Sci. USA 111, 7385–7390 (2014).
Weng, D. et al. Caspase-8 and RIP kinases regulate bacteria-induced innate immune responses and cell death. Proc. Natl. Acad. Sci. USA 111, 7391–7396 (2014).
Lukens, J.R. et al. RIP1-driven autoinflammation targets IL-1α independently of inflammasomes and RIP3. Nature 498, 224–227 (2013).
Takahashi, N. et al. Necrostatin-1 analogues: critical issues on the specificity, activity and in vivo use in experimental disease models. Cell Death Dis. 3, e437 (2012).
Sun, L. et al. Mixed lineage kinase domain-like protein mediates necrosis signaling downstream of RIP3 kinase. Cell 148, 213–227 (2012).
Wang, Z., Jiang, H., Chen, S., Du, F. & Wang, X. The mitochondrial phosphatase PGAM5 functions at the convergence point of multiple necrotic death pathways. Cell 148, 228–243 (2012).
Ichinohe, T., Yamazaki, T., Koshiba, T. & Yanagi, Y. Mitochondrial protein mitofusin 2 is required for NLRP3 inflammasome activation after RNA virus infection. Proc. Natl. Acad. Sci. USA 110, 17963–17968 (2013).
Shimada, K. et al. Oxidized mitochondrial DNA activates the NLRP3 inflammasome during apoptosis. Immunity 36, 401–414 (2012).
Nakahira, K. et al. Autophagy proteins regulate innate immune responses by inhibiting the release of mitochondrial DNA mediated by the NALP3 inflammasome. Nat. Immunol. 12, 222–230 (2011).
Lupfer, C. et al. Receptor interacting protein kinase 2-mediated mitophagy regulates inflammasome activation during virus infection. Nat. Immunol. 14, 480–488 (2013).
Stout-Delgado, H.W., Vaughan, S.E., Shirali, A.C., Jaramillo, R.J. & Harrod, K.S. Impaired NLRP3 inflammasome function in elderly mice during influenza infection is rescued by treatment with nigericin. J. Immunol. 188, 2815–2824 (2012).
Knott, A.B., Perkins, G., Schwarzenbacher, R. & Bossy-Wetzel, E. Mitochondrial fragmentation in neurodegeneration. Nat. Rev. Neurosci. 9, 505–518 (2008).
Losón, O.C., Song, Z., Chen, H. & Chan, D.C. Fis1, Mff, MiD49, and MiD51 mediate Drp1 recruitment in mitochondrial fission. Mol. Biol. Cell 24, 659–667 (2013).
Ishihara, N. et al. Mitochondrial fission factor Drp1 is essential for embryonic development and synapse formation in mice. Nat. Cell Biol. 11, 958–966 (2009).
Poeck, H. et al. Recognition of RNA virus by RIG-I results in activation of CARD9 and inflammasome signaling for interleukin 1β production. Nat. Immunol. 11, 63–69 (2010).
Mitoma, H. et al. The DHX33 RNA helicase senses cytosolic RNA and activates the NLRP3 inflammasome. Immunity 39, 123–135 (2013).
Janeway, C.A. Jr. & Medzhitov, R. Innate immune recognition. Annu. Rev. Immunol. 20, 197–216 (2002).
Ryu, S.W., Jeong, H.J., Choi, M., Karbowski, M. & Choi, C. Optic atrophy 3 as a protein of the mitochondrial outer membrane induces mitochondrial fragmentation. Cell. Mol. Life Sci. 67, 2839–2850 (2010).
Moujalled, D.M., Cook, W.D., Murphy, J.M. & Vaux, D.L. Necroptosis induced by RIPK3 requires MLKL but not Drp1. Cell Death Dis. 5, e1086 (2014).
Martinon, F., Petrilli, V., Mayor, A., Tardivel, A. & Tschopp, J. Gout-associated uric acid crystals activate the NALP3 inflammasome. Nature 440, 237–241 (2006).
Newton, K., Sun, X. & Dixit, V.M. Kinase RIP3 is dispensable for normal NF-κBs, signaling by the B-cell and T-cell receptors, tumor necrosis factor receptor 1, and Toll-like receptors 2 and 4. Mol. Cell. Biol. 24, 1464–1469 (2004).
Kato, H. et al. Cell type-specific involvement of RIG-I in antiviral response. Immunity 23, 19–28 (2005).
Gitlin, L. et al. Essential role of mda-5 in type I IFN responses to polyriboinosinic:polyribocytidylic acid and encephalomyocarditis picornavirus. Proc. Natl. Acad. Sci. USA 103, 8459–8464 (2006).
Wu, J. et al. Mlkl knockout mice demonstrate the indispensable role of Mlkl in necroptosis. Cell Res. 23, 994–1006 (2013).
Alexopoulou, L., Holt, A.C., Medzhitov, R. & Flavell, R.A. Recognition of double-stranded RNA and activation of NF-κB by Toll-like receptor 3. Nature 413, 732–738 (2001).
Yan, Y. et al. Omega-3 fatty acids prevent inflammation and metabolic disorder through inhibition of NLRP3 inflammasome activation. Immunity 38, 1154–1163 (2013).
Acknowledgements
We thank J. Tschopp (University of Lausanne) for Nlrp3−/− mice; S. Akira (Osaka University) for Rig-I−/− mice; V.M. Dixit (Genentech) for Rip3−/− mice; M. Colonna (Washington University) for Mda5−/− mice; Z. Jiang (Peking University) for Sendai virus and VSV (Indiana strain); and Z. Song (Wuhan University) for the DRP1 construct. Supported by the National Basic Research Program of China (2014CB910800), the National Natural Science Foundation of China (81330078 and 81222040), the Doctoral Fund of the Ministry of Education of China (20123402120001, 20123402110010), the One Hundred Person Project (R.Z.) and the Fundamental Research Funds for the Central Universities (R.Z. and W.J.).
Author information
Authors and Affiliations
Contributions
X.W., W.J., Y.Y. and T.G. performed the experiments; W.J., J.H., Z.T. and R.Z. designed the research; X.W., W.J., Z.T. and R.Z. wrote the manuscript; and R.Z. and Z.T. supervised the project.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Integrated supplementary information
Supplementary Figure 1 Role of RIP1-RIP3 and PGAM5 in RNA virus–induced inflammasome activation.
(a) LDH release from LPS-primed BMDMs from wild-type mice (WT), Rip3-/-or Nlrp3-/-mice infected with VSV for 6 hours or stimulated with nigericin for 30 min. (b) qPCR analysis of Ifn-β expression in non-primed BMDMs from WT, Rip3-/- or Nlrp3-/- mice infected with VSV as indicated doses for 6 hours. (c) Immunoblot analysis of RIP1 expression in BMDMs transfected with siRNA against Rip1 for 48 hours. (d) Immunoblot analysis of pro-IL-1β or NLRP3 expression in BMDMs transfected with siRNA against Rip1 and stimulated with Pam3CSK4 for 6 hours. (e) Immunoblot analysis of PGAM5 in THP-1 cells stably expressing shRNA against PGAM5. (f, g) IL-1β secretion from THP-1 cells stably expressing shRNA against PGAM5 infected with VSV for 6 hours (f), or stimulated with MSU for 4 hours or nigericin for 30 min (g). (h) Confocal microscopy analysis in LPS-primed BMDMs from Rip3+/+ or Rip3-/-mice infected with VSV for 2 hours or stimulated with nigericin for 30 min, and then stained with MitoSOX and DAPI. Scale bar, 50 μm. (i) Confocal microscopy analysis in LPS-primed BMDMs pretreated with Nec-1s (30 μM) for 1 hour and then infected with VSV for 2 hours or stimulated with nigericin for 30 min. After treatment, the cells were stained with MitoSOX and DAPI. Scale bar, 50 μm. NS, P > 0.05 (two-way ANOVA (a,b,f,g). Data are representative of at three independent experiments (c-e, h, i) or are from three independent experiments (with biological duplicates in each) (a, b, f, g; mean and s.e.m.).
Supplementary Figure 2 RIP3 is required for VSV-induced translocation of ASC to mitochondria.
Confocal microscopy analysis in LPS-primed BMDMs from Rip3+/+ or Rip3-/- mice infected with VSV for 2 hours and then stained with anti-ASC antibody, Mitotracker red and DAPI. Data are representative of at three independent experiments.
Supplementary Figure 3 RIP3 is required for VSV-induced translocation of DRP1 to mitochondria.
Confocal microscopy analysis in LPS-primed BMDMs from Rip3+/+ or Rip3-/- mice infected with VSV for 2 hours or stimulated with nigericin for 30 min, and then stained with anti-DRP1 antibody, Mitotracker and DAPI. Scale bar, 20 μm. Data are representative of at three independent experiments.
Supplementary Figure 4 RIP1 is required for VSV-induced translocation of DRP1 to mitochondria.
Confocal microscopy analysis in LPS-primed BMDMs pretreated with Nec-1s (30 μM) for 1 hour and then were infected with VSV for 2 hours or stimulated with nigericin for 30 min. After treatment, the cells were stained with anti-DRP1 antibody, Mitotracker and DAPI. Scale bar, 20 μm. Data are representative of at three independent experiments
Supplementary Figure 5 MLKL is not required for VSV-induced translocation of DRP1 to mitochondria.
Confocal microscopy analysis in LPS-primed BMDMs from Mlkl+/+ or Mlkl-/- mice were infected with VSV for 2 hours or stimulated with nigericin for 30 min, and then stained with anti-DRP1 antibody and Mitotracker. Scale bar, 20 μm. Data are representative of at three independent experiments.
Supplementary Figure 6 Role of DRP1 or FIS1 in mitochondrial damage and inflammasome activation.
(a) Immunoblot analysis of BMDMs transfected with siRNA against Drp1 for 48 hours. (b) Confocal microscopy analysis of LPS-primed BMDMs transfected with siRNA against Drp1 and then infected with VSV for 2 hours or stimulated with nigericin for 30 min and followed by staining with MitoSOX and DAPI. Scale bar, 50 μm. (c) IL-1β secretion from LPS-primed BMDMs transfected with siRNA against Drp1 and then stimulated with MSU, poly (A:T) for 4 hours or ATP, nigericin for 30 min. (d) Immunoblot analysis of BMDMs transfected with siRNA against Fis1 for 48 hours. (e, f) Confocal microscopy analysis of LPS-primed BMDMs transfected with siRNA against Fis1 and infected with VSV for 2 hours or stimulated with nigericin for 30 min and followed by staining with Mitotracker red. Representative pictures (e) or qualification (f) was shown. Scale bar, 50 μm. * P < 0.001, NS P > 0.05. Data are representative of at three independent experiments (a, b, d, e) or are from three independent experiments (with biological duplicates in each) (c, f; mean and s.e.m.).
Supplementary Figure 7 Role of RIP3 and DRP1 in CCCP-induced mitochondrial fission and IL-1β production.
(a) Immunoblot analysis of DRP1 phosphorylation in LPS-primed BMDMs stimulated with CCCP (10 μM) or infected with VSV for 1 hours. (b) IL-1β secretion from LPS-primed BMDMs from Rip3+/+ or Rip3-/-mice were stimulated with CCCP for 5 hours. (c) Confocal microscopy analysis of LPS-primed BMDMs from Rip3+/+ or Rip3-/- mice stimulated with CCCP (10 μM) for 2 hours, and then stained with anti-DRP1 antibody and Mitotracker. Scale bar, 25 μm. (d) IL-1β secretion from LPS-primed BMDMs transfected with siRNA against Drp1 and then stimulated with CCCP (10 μM) for 5 hours. (e, f) Confocal microscopy analysis of LPS-primed BMDMs transfected with siRNA against Drp1 and stimulated with CCCP (10 μM) for 2 hours, and followed by staining with Mitotracker red and DAPI. Representative pictures (e) or qualification (f) was shown. Scale bar, 25 μm. NS P > 0.05, * P<0.01 (two-way ANOVA), * P<0.001 (two-way ANOVA). Data are representative of at three independent experiments (a, c, e) or are from three independent experiments (with biological duplicates in each) (b, d, f; mean and s.e.m.).
Supplementary Figure 8 Role of RNA sensors in VSV- or dsRNA-induced activation of the NLRP3 inflammasome.
(a, b) IL-1β secretion from LPS-primed BMDMs from wild-type mice, Rig-I-/- or Mda5-/- mice infected with VSV (a) or transfected with poly (I:C) (b) for 6 hours. (c) qPCR analysis of Ifn-β expression in non-primed BMDMs from wild-type mice, Rig-I-/- or Mda5-/- mice infected with VSV for 6 hours. (d) Immunoblot analysis of THP-1 cells stably expressing shRNA against Rip1 or DHX33. (e) IL-1β secretion from THP-1 cells stably expressing shRNA against Rip1 or DHX33 infected with VSV for 6 hours or stimulated with nigericin for 30 min. (f, g) IL-1β secretion from Pam3CSK4-primed BMDMs from Tlr3+/+ or Tlr3-/-mice infected with VSV (f) or stimulated with poly (I:C) transfection (g) for 6 hours. NS P > 0.05 (unpaired t-test (a, b, c, g), NS P > 0.05 (two-way ANOVA (f)), * P < 0.01 (two-way ANOVA (e)), ** P < 0.001 (two-way ANOVA (c)). Data are representative of at three independent experiments (d) or are from three independent experiments (with biological duplicates in each) (a-c, e-g; mean and s.e.m.).
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–8 and Supplementary Table 1 (PDF 5859 kb)
Rights and permissions
About this article
Cite this article
Wang, X., Jiang, W., Yan, Y. et al. RNA viruses promote activation of the NLRP3 inflammasome through a RIP1-RIP3-DRP1 signaling pathway. Nat Immunol 15, 1126–1133 (2014). https://doi.org/10.1038/ni.3015
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ni.3015
This article is cited by
-
The role of cell death in SARS-CoV-2 infection
Signal Transduction and Targeted Therapy (2023)
-
A necroptosis-independent function of RIPK3 promotes immune dysfunction and prevents control of chronic LCMV infection
Cell Death & Disease (2023)
-
Mitophagy for cardioprotection
Basic Research in Cardiology (2023)
-
An overview on in vitro and in vivo antiviral activity of lactoferrin: its efficacy against SARS-CoV-2 infection
BioMetals (2023)
-
Interleukin-1 alpha and high mobility group box-1 secretion in polyinosinic:polycytidylic-induced colorectal cancer cells occur via RIPK1-dependent mechanism and participate in tumourigenesis
Journal of Cell Communication and Signaling (2023)