Abstract
The mammalian intestinal tract harbors a diverse community of trillions of microorganisms, which have co-evolved with the host immune system for millions of years. Many of these microorganisms perform functions critical for host physiology, but the host must remain vigilant to control the microbial community so that the symbiotic nature of the relationship is maintained. To facilitate homeostasis, the immune system ensures that the diverse microbial load is tolerated and anatomically contained, while remaining responsive to microbial breaches and invasion. Although the microbiota is required for intestinal immune development, immune responses also regulate the structure and composition of the intestinal microbiota. Here we discuss recent advances in our understanding of these complex interactions and their implications for human health and disease.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
Change history
20 September 2013
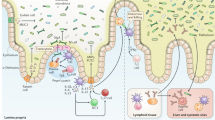
In the version of this article initially published, a label was missing from Figure 2. The lymphoid structure in the large intestine should be labeled 'Isolated lymphoid follicle'. The error has been corrected in the HTML and PDF versions of the article.
References
Xu, J. & Gordon, J.I. Honor thy symbionts. Proc. Natl. Acad. Sci. USA 100, 10452–10459 (2003).
Eckburg, P.B. et al. Diversity of the human intestinal microbial flora. Science 308, 1635–1638 (2005).This is the first comprehensive study to use a culture-independent approach to describe the composition of the intestinal microbiota in healthy adult humans.
Ley, R.E. et al. Evolution of mammals and their gut microbes. Science 320, 1647–1651 (2008).
Human Microbiome Project Consortium. Structure, function and diversity of the healthy human microbiome. Nature 486, 207–214 (2012).
Sekirov, I., Russel, S.L., Antunes, L.C.M. & Finlay, B.B. Gut microbiota in health and disease. Physiol. Rev. 90, 859–904 (2010).
Garrett, W.S., Gordon, J.I. & Glimcher, L.H. Homeostasis and inflammation in the intestine. Cell 140, 859–870 (2010).
Willing, B.P., Russell, S.L. & Finlay, B.B. Shifting the balance: antibiotic effects on host-microbiota mutualism. Nat. Rev. Microbiol. 9, 233–243 (2011).
Maslowski, K.M. & Mackay, C.R. Diet, gut microbiota and immune responses. Nat. Immunol. 12, 5–9 (2011).
Gill, N., Wlodarska, M. & Finlay, B.B. Roadblocks in the gut: barriers to enteric infection. Cell. Microbiol. 13, 660–669 (2011).
Willing, B.P., Gill, N. & Finlay, B.B. The role of the immune system in regulating the microbiota. Gut Microbes 1, 213–223 (2010).
Hooper, L.V. & Macpherson, A.J. Immune adaptations that maintain homeostasis with the intestinal microbiota. Nat. Rev. Immunol. 10, 159–169 (2010).
Kim, Y.S. & Ho, S.B. Intestinal goblet cells and mucins in health and disease: recent insights and progress. Curr. Gastroenterol. Rep. 12, 319–330 (2010).
Johansson, M.E. et al. The inner of the two Muc2 mucin-dependent mucus layers in colon is devoid of bacteria. Proc. Natl. Acad. Sci. USA 105, 15064–15069 (2008).This study provides the first visual evidence of the composition of the mucus layer, highlighting the function of the mucus layer in segregating the microbiota away from the host epithelium.
Wlodarska, M. et al. Antibiotic treatment alters the colonic mucus layer and predisposes the host to exacerbated Citrobacter rodentium–induced colitis. Infect. Immun. 79, 1536–1545 (2011).
Fyderek, K. Mucosal bacterial microflora and mucus layer thickness in adolescents with inflammatory bowel disease. World J. Gastroenterol. 15, 5287 (2009).
Johansson, M., Larsson, J. & Hansson, G. The two mucus layers of colon are organized by the MUC2 mucin, whereas the outer layer is a legislator of host-microbial interactions. Proc. Natl. Acad. Sci. USA 108 (suppl. 1), 4659–4665 (2011).
Podolsky, D.K. et al. Identification of human intestinal trefoil factor. Goblet cell-specific expression of a peptide targeted for apical secretion. J. Biol. Chem. 268, 6694–6702 (1993).
Artis, D. et al. RELMbeta/FIZZ2 is a goblet cell-specific immune-effector molecule in the gastrointestinal tract. Proc. Natl. Acad. Sci. USA 101, 13596–13600 (2004).
Bevins, C.L. & Salzman, N.H. Paneth cells, antimicrobial peptides and maintenance of intestinal homeostasis. Nat. Rev. Microbiol. 9, 356–368 (2011).
Vaishnava, S., Behrendt, C.L., Ismail, A.S., Eckmann, L. & Hooper, L.V. Paneth cells directly sense gut commensals and maintain homeostasis at the intestinal host-microbial interface. Proc. Natl. Acad. Sci. USA 105, 20858–20863 (2008).
Vaishnava, S. et al. The antibacterial lectin RegIIIgamma promotes the spatial segregation of microbiota and host in the intestine. Science 334, 255–258 (2011).
Selsted, M.E. & Ouellette, A.J. Mammalian defensins in the antimicrobial immune response. Nat. Immunol. 6, 551–557 (2005).
Salzman, N. et al. Enteric defensins are essential regulators of intestinal microbial ecology. Nat. Immunol. 11, 76–83 (2010).
Brandl, K., Plitas, G., Schnabl, B., DeMatteo, R.P. & Pamer, E.G. MyD88-mediated signals induce the bactericidal lectin RegIII gamma and protect mice against intestinal Listeria monocytogenes infection. J. Exp. Med. 204, 1891–1900 (2007).
Kaiser, V. & Diamond, G. Expression of mammalian defensin genes. J. Leukoc. Biol. 68, 779–784 (2000).
Menendez, A. et al. Bacterial stimulation of the TLR-MyD88 pathway modulates the homeostatic expression of ileal Paneth cell α-defensins. J. Innate Immun. 5, 39–49 (2013).
Chu, H. et al. Human alpha-defensin 6 promotes mucosal innate immunity through self-assembled peptide nanonets. Science 337, 477–481 (2012).
Schroeder, B. et al. Reduction of disulphide bonds unmasks potent antimicrobial activity of human β-defensin 1. Nature 469, 419–423 (2011).
Spits, H. & Di Santo, J.P. The expanding family of innate lymphoid cells: regulators and effectors of immunity and tissue remodeling. Nat. Immunol. 12, 21–27 (2011).
Spits, H. & Cupedo, T. Innate lymphoid cells: emerging insights in development, lineage relationships, and function. Annu. Rev. Immunol. 30, 647–675 (2012).
Sonnenberg, G. & Artis, D. Innate lymphoid cell interactions with microbiota: implications for intestinal health and disease. Immunity 37, 601–610 (2012).
Qiu, J. et al. The aryl hydrocarbon receptor regulates gut immunity through modulation of innate lymphoid cells. Immunity 36, 92–104 (2012).
Sawa, S. et al. RORgammat+ innate lymphoid cells regulate intestinal homeostasis by integrating negative signals from the symbiotic microbiota. Nat. Immunol. 12, 320–326 (2011).
Lochner, M. et al. Microbiota-induced tertiary lymphoid tissues aggravate inflammatory disease in the absence of RORgamma t and LTi cells. J. Exp. Med. 208, 125–134 (2011).
Sonnenberg, G. et al. Innate lymphoid cells promote anatomical containment of lymphoid-resident commensal bacteria. Science 336, 1321–1325 (2012).
Sonnenberg, G.F., Monticelli, L.A., Elloso, M.M., Fouser, L.A. & Artis, D. CD4+ lymphoid tissue-inducer cells promote innate immunity in the gut. Immunity 34, 122–134 (2011).
Monticelli, L.A. et al. Innate lymphoid cells promote lung-tissue homeostasis after infection with influenza virus. Nat. Immunol. 12, 1045–1054 (2011).
Powell, N. et al. The transcription factor T-bet regulates intestinal inflammation mediated by interleukin-7 receptor+ innate lymphoid cells. Immunity 37, 674–684 (2012).
Cebra, J.J. Influences of microbiota on intestinal immune system development. Am. J. Clin. Nutr. 69, 1046S–1051S (1999).
Abreu, M.T. Toll-like receptor signalling in the intestinal epithelium: how bacterial recognition shapes intestinal function. Nat. Rev. Immunol. 10, 131–144 (2010).
Takeuchi, O. & Akira, S. Pattern recognition receptors and inflammation. Cell 140, 805–820 (2010).
Carvalho, F.A., Aitken, J.D., Vijay-Kumar, M. & Gewirtz, A.T. Toll-like receptor-gut microbiota interactions: perturb at your own risk!. Annu. Rev. Physiol. 74, 177–198 (2012).
Ubeda, C. et al. Familial transmission rather than defective innate immunity shapes the distinct intestinal microbiota of TLR-deficient mice. J. Exp. Med. 209, 1445–1456 (2012).
Vijay-Kumar, M. et al. Metabolic syndrome and altered gut microbiota in mice lacking Toll-like receptor 5. Science 328, 228–231 (2010).
Vijay-Kumar, M. et al. Deletion of TLR5 results in spontaneous colitis in mice. J. Clin. Invest. 117, 3909–3921 (2007).
Slack, E. et al. Innate and adaptive immunity cooperate flexibly to maintain host-microbiota mutualism. Science 325, 617–620 (2009).
Salcedo, R. et al. MyD88-mediated signaling prevents development of adenocarcinomas of the colon: role of interleukin 18. J. Exp. Med. 207, 1625–1636 (2010).
Elinav, E. et al. NLRP6 inflammasome regulates colonic microbial ecology and risk for colitis. Cell 145, 745–757 (2011).This work demonstrates that both the microbiota and the host's genotype can affect mucosal disease, and disease susceptibility can be transferred to another wild-type host by a colitogenic microbiota.
Huber, S. et al. IL-22BP is regulated by the inflammasome and modulates tumorigenesis in the intestine. Nature 491, 259–263 (2012).
Santaolalla, R. & Abreu, M.T. Innate immunity in the small intestine. Curr. Opin. Gastroenterol. 28, 124–129 (2012).
Coombes, J.L. & Powrie, F. Dendritic cells in intestinal immune regulation. Nat. Rev. Immunol. 8, 435–446 (2008).
Farache, J. et al. Luminal bacteria recruit CD103+ dendritic cells into the intestinal epithelium to sample bacterial antigens for presentation. Immunity 38, 581–595 (2013).
McDole, J.R. et al. Goblet cells deliver luminal antigen to CD103+ dendritic cells in the small intestine. Nature 483, 345–349 (2012).
Knoop, K.A., Miller, M.J. & Newberry, R.D. Transepithelial antigen delivery in the small intestine: different paths, different outcomes. Curr. Opin. Gastroenterol. 29, 112–118 (2013).
Mestecky, J. & Russell, M.W. Specific antibody activity, glycan heterogeneity and polyreactivity contribute to the protective activity of S-IgA at mucosal surfaces. Immunol. Lett. 124, 57–62 (2009).
Niess, J.H. et al. CX3CR1-mediated dendritic cell access to the intestinal lumen and bacterial clearance. Science 307, 254–258 (2005).
Macpherson, A.J. & Uhr, T. Induction of protective IgA by intestinal dendritic cells carrying commensal bacteria. Science 303, 1662–1665 (2004).This report demonstrates that specialized bacteria-laden dendritic cells can induce protective IgA to protect the host epithelium from bacterial invasion, and migration of these dendritic cells is limited to the mesenteric lymph nodes of the mucosal immune system.
Hapfelmeier, S. et al. Reversible microbial colonization of germ-free mice reveals the dynamics of IgA immune responses. Science 328, 1705–1709 (2010).
Peterson, D.A., McNulty, N.P., Guruge, J.L. & Gordon, J.I. IgA response to symbiotic bacteria as a mediator of gut homeostasis. Cell Host Microbe 2, 328–339 (2007).
Macpherson, A.J. et al. A primitive T cell–independent mechanism of intestinal mucosal IgA responses to commensal bacteria. Science 288, 2222–2226 (2000).
Suzuki, K. et al. Aberrant expansion of segmented filamentous bacteria in IgA-deficient gut. Proc. Natl. Acad. Sci. USA 101, 1981–1986 (2004).
Wei, M. et al. Mice carrying a knock-in mutation of Aicda resulting in a defect in somatic hypermutation have impaired gut homeostasis and compromised mucosal defense. Nat. Immunol. 12, 264–270 (2011).
Slack, E., Balmer, M.L., Fritz, J.H. & Hapfelmeier, S. Functional flexibility of intestinal IgA—broadening the fine line. Front. Immunol. 3, 100 (2012).
Cong, Y., Feng, T., Fujihashi, K., Schoeb, T. & Elson, C. A dominant, coordinated T regulatory cell-IgA response to the intestinal microbiota. Proc. Natl. Acad. Sci. USA 106, 19256–19261 (2009).This study extends the role for T reg cells to include the induction and maintainance of IgA+ plasma cells in the intestine, and promotion of mutualism with the microbiota.
Read, S., Malmstrom, V. & Powrie, F. Cytotoxic T lymphocyte–associated antigen 4 plays an essential role in the function of CD25+CD4+ regulatory cells that control intestinal inflammation. J. Exp. Med. 192, 295–302 (2000).
Maloy, K.J. et al. CD4+CD25+ T(R) cells suppress innate immune pathology through cytokine-dependent mechanisms. J. Exp. Med. 197, 111–119 (2003).
Li, M.O., Wan, Y.Y. & Flavell, R.A. T cell-produced transforming growth factor-β1 controls T cell tolerance and regulates TH1- and TH17-cell differentiation. Immunity 26, 579–591 (2007).
Ahern, P.P. et al. Interleukin-23 drives intestinal inflammation through direct activity on T cells. Immunity 33, 279–288 (2010).
Littman, D.R. & Rudensky, A.Y. Th17 and regulatory T cells in mediating and restraining inflammation. Cell 140, 845–858 (2010).
Chaudhry, A. et al. CD4+ regulatory T cells control TH17 responses in a Stat3-dependent manner. Science 326, 986–991 (2009).
Maynard, C.L. et al. Regulatory T cells expressing interleukin 10 develop from Foxp3+ and Foxp3− precursor cells in the absence of interleukin 10. Nat. Immunol. 8, 931–941 (2007).
Foussat, A. et al. A comparative study between T regulatory type 1 and CD4+CD25+ T cells in the control of inflammation. J. Immunol. 171, 5018–5026 (2003).
Lupp, C. et al. Host-mediated inflammation disrupts the intestinal microbiota and promotes the overgrowth of Enterobacteriaceae. Cell Host Microbe 2, 119–129 (2007).This study reveals that the composition of the intestinal microbiota changes in distinctive ways in response to infection and inflammation, and underscores the importance of intestinal microbial ecosystems during infection.
Littman, D.R. & Pamer, E.G. Role of the commensal microbiota in normal and pathogenic host immune responses. Cell Host Microbe 10, 311–323 (2011).
Stecher, B. et al. Salmonella enterica serovar typhimurium exploits inflammation to compete with the intestinal microbiota. PLoS Biol. 5, 2177–2189 (2007).
Winter, S.E. et al. Gut inflammation provides a respiratory electron acceptor for Salmonella. Nature 467, 426–429 (2010).
Winter, S.E. et al. Host-derived nitrate boosts growth of E. coli in the inflamed gut. Science 339, 708–711 (2013).
Gill, N. et al. Neutrophil elastase alters the murine gut microbiota resulting in enhanced Salmonella colonization. PLoS ONE 7, e49646 (2012).
Stelter, C. et al. Salmonella-induced mucosal lectin RegIIIb kills competing gut microbiota. PLoS ONE 6, e20749 (2011).
Raetz, M. et al. Parasite-induced TH1 cells and intestinal dysbiosis cooperate in IFN-gamma–dependent elimination of Paneth cells. Nat. Immunol. 14, 136–142 (2013).
Khor, B., Gardet, A. & Xavier, R.J. Genetics and pathogenesis of inflammatory bowel disease. Nature 474, 307–317 (2011).
Jostins, L. et al. Host-microbe interactions have shaped the genetic architecture of inflammatory bowel disease. Nature 491, 119–124 (2012).
Wehkamp, J. et al. NOD2 (CARD15) mutations in Crohn's disease are associated with diminished mucosal alpha-defensin expression. Gut 53, 1658–1664 (2004).
Ogura, Y. et al. A frameshift mutation in NOD2 associated with susceptibity to Crohn's disease. Nature 411, 603–606 (2001).
Hugot, J.P. et al. Association of NOD-2 leucine-rich repeat variants with susceptibility to Crohn's disease. Nature 411, 599–603 (2001)References 84 and 85 reported NOD2 as a susceptibility locus for Crohn's disease, providing evidence the first genetic link to IBD and insight into how a dysregulated immune response to the microbiota can lead to inflammatory diseases.
Petnicki-Ocwieja, T. et al. Nod2 is required for the regulation of commensal microbiota in the intestine. Proc. Natl. Acad. Sci. USA 106, 15813–15818 (2009).
Kobayashi, K.S. et al. Nod2-dependent regulation of innate and adaptive immunity in the intestinal tract. Science 307, 731–734 (2005).
Wehkamp, J. et al. Reduced Paneth cell alpha-defensins in ileal Crohn's disease. Proc. Natl. Acad. Sci. USA 102, 18129–18134 (2005).
Simms, L.A. et al. Reduced alpha-defensin expression is associated with inflammation and not NOD2 mutation status in ileal Crohn's disease. Gut 57, 903–910 (2008).
Bernink, J.H. et al. Human type 1 innate lymphoid cells accumulate in inflamed mucosal tissues. Nat. Immunol. 14, 221–229 (2013).
Mehandru, S. et al. Mechanisms of gastrointestinal CD4+ T-cell depletion during acute and early human immunodeficiency virus type 1 infection. J. Virol. 81, 599–612 (2007).
Brenchley, J.M. et al. Differential TH17 CD4 T-cell depletion in pathogenic and nonpathogenic lentiviral infections. Blood 112, 2826–2835 (2008).
Gosselin, A. et al. Peripheral blood CCR4+CCR6+ and CXCR3+CCR6+CD4+ T cells are highly permissive to HIV-1 infection. J. Immunol. 184, 1604–1616 (2010).
Brenchley, J.M. & Douek, D.C. HIV infection and the gastrointestinal immune system. Mucosal Immunol. 1, 23–30 (2008).
Raffatellu, M. et al. Simian immunodeficiency virus-induced mucosal interleukin-17 deficiency promotes Salmonella dissemination from the gut. Nat. Med. 14, 421–428 (2008).
Macal, M. et al. Effective CD4+ T-cell restoration in gut-associated lymphoid tissue of HIV-infected patients is associated with enhanced Th17 cells and polyfunctional HIV-specific T-cell responses. Mucosal Immunol. 1, 475–488 (2008).
Cecchinato, V. et al. Altered balance between Th17 and Th1 cells at mucosal sites predicts AIDS progression in simian immunodeficiency virus-infected macaques. Mucosal Immunol. 1, 279–288 (2008).
Favre, D. et al. Critical loss of the balance between Th17 and T regulatory cell populations in pathogenic SIV infection. PLoS Pathog. 5, e1000295 (2009).
Saxena, D. et al. Human microbiome and HIV/AIDS. Curr. HIV/AIDS Rep. 9, 44–51 (2012).
Ellis, C.L. et al. Molecular characterization of stool microbiota in HIV-infected subjects by panbacterial and order-level 16S ribosomal DNA (rDNA) quantification and correlations with immune activation. J. Acquir. Immune Defic. Syndr. 57, 363–370 (2011).
Malamut, G. et al. The enteropathy associated with common variable immunodeficiency: the delineated frontiers with celiac disease. Am. J. Gastroenterol. 105, 2262–2275 (2010).
Shulzhenko, N. et al. Crosstalk between B lymphocytes, microbiota and the intestinal epithelium governs immunity versus metabolism in the gut. Nat. Med. 17, 1585–1593 (2011).
Mannon, P.J. et al. Excess IL-12 but not IL-23 accompanies the inflammatory bowel disease associated with common variable immunodeficiency. Gastroenterology 131, 748–756 (2006).
Scamurra, R.W. et al. Mucosal plasma cell repetoire during HIV-1 infection. J. Immunol. 169, 4008–4016 (2002).
Man, S.M., Kaakoush, N.O. & Mitchell, H.M. The role of bacteria and pattern-recognition receptors in Crohn's disease. Nat. Rev. Gastroenterol. Hepatol. 8, 152–168 (2011).
Duerkop, B.A. & Hooper, L.V. Resident viruses and their interactions with the immune system. Nat. Immunol. (18 Jun 2013) doi:10.1038/ni.2614.
Acknowledgements
We thank M. Wlodarska, L. Reynolds and N. Gill for the critical revision of this manuscript and thoughtful insights. The Finlay laboratory is supported by operating grants from Canadian Institutes of Health Research.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Rights and permissions
About this article
Cite this article
Brown, E., Sadarangani, M. & Finlay, B. The role of the immune system in governing host-microbe interactions in the intestine. Nat Immunol 14, 660–667 (2013). https://doi.org/10.1038/ni.2611
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ni.2611
This article is cited by
-
Effects of transport stress on the oxidative index, apoptosis and autophagy in the small intestine of caprine
BMC Veterinary Research (2023)
-
Gut microbiota in cancer: insights on microbial metabolites and therapeutic strategies
Medical Oncology (2023)
-
Oral administration of asparagine and 3-indolepropionic acid prolongs survival time of rats with traumatic colon injury
Military Medical Research (2022)
-
RNA-Sequencing of Heterorhabditis nematodes to identify factors involved in symbiosis with Photorhabdus bacteria
BMC Genomics (2022)
-
Impacts and mechanisms of metabolic reprogramming of tumor microenvironment for immunotherapy in gastric cancer
Cell Death & Disease (2022)