Abstract
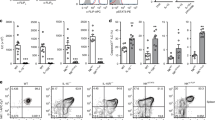
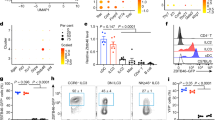
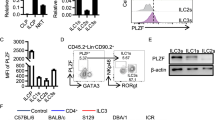
NKp46+ innate lymphoid cells (ILCs) serve important roles in regulating the intestinal microbiota and defense against pathogens. Whether NKp46+ ILCs arise directly from lymphoid tissue–inducer (LTi) cells or represent a separate lineage remains controversial. We report here that the transcription factor T-bet (encoded by Tbx21) was essential for the development of NKp46+ ILCs but not of LTi cells or nuocytes. Deficiency in interleukin 22 (IL-22)-producing NKp46+ ILCs resulted in greater susceptibility of Tbx21−/− mice to intestinal infection. Haploinsufficient T-bet expression resulted in lower expression of the signaling molecule Notch, and Notch signaling was necessary for the transition of LTi cells into NKp46+ ILCs. Furthermore, NKp46+ ILCs differentiated solely from the CD4− LTi population, not the CD4+ LTi population. Our results pinpoint the regulation of Notch signaling by T-bet as a distinct molecular pathway that guides the development of NKp46+ ILCs.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
Accession codes
Change history
21 March 2013
In the version of this article initially published, the key to Figure 1c was labeled incorrectly. The correct labeling is as follows: dotted line, Isotype; shaded box, T-bet. The error has been corrected in the HTML and PDF versions of the article.
References
Sonnenberg, G.F., Fouser, L.A. & Artis, D. Border patrol: regulation of immunity, inflammation and tissue homeostasis at barrier surfaces by IL-22. Nat. Immunol. 12, 383–390 (2011).
Sanos, S.L., Vonarbourg, C., Mortha, A. & Diefenbach, A. Control of epithelial cell function by interleukin-22-producing RORgammat+ innate lymphoid cells. Immunology 132, 453–465 (2011).
Moro, K. et al. Innate production of TH2 cytokines by adipose tissue-associated c-Kit+Sca-1+ lymphoid cells. Nature 463, 540–544 (2009).
Neill, D.R. et al. Nuocytes represent a new innate effector leukocyte that mediates type-2 immunity. Nature 464, 1367–1370 (2010).
Wong, S.H. et al. Transcription factor RORα is critical for nuocyte development. Nat. Immunol. 13, 229–236 (2012).
Spits, H. & Di Santo, J.P. The expanding family of innate lymphoid cells: regulators and effectors of immunity and tissue remodeling. Nat. Immunol. 12, 21–27 (2011).
Eberl, G. et al. An essential function for the nuclear receptor Rorγt in the generation of fetal lymphoid tissue inducer cells. Nat. Immunol. 5, 64–73 (2004).
Sawa, S. et al. Lineage relationship analysis of RORγt+ innate lymphoid cells. Science 330, 665–669 (2010).
Yokota, Y. et al. Development of peripheral lymphoid organs and natural killer cells depends on the helix-loop-helix inhibitor Id2. Nature 397, 702–706 (1999).
Gascoyne, D.M. et al. The basic leucine zipper transcription factor E4BP4 is essential for natural killer cell development. Nat. Immunol. 10, 1118–1124 (2009).
Kamizono, S. et al. Nfil3/E4bp4 is required for the development and maturation of NK cells in vivo. J. Exp. Med. 206, 2977–2986 (2009).
Hoyler, T. et al. The transcription factor GATA-3 controls cell fate and maintenance of type 2 innate lymphoid cells. Immunity 37, 634–648 (2012).
Eberl, G. Development and evolution of RORγt+ cells in a microbe's world. Immunol. Rev. 245, 177–188 (2012).
Bouskra, D. et al. Lymphoid tissue genesis induced by commensals through NOD1 regulates intestinal homeostasis. Nature 456, 507–510 (2008).
Cupedo, T. et al. Human fetal lymphoid tissue–inducer cells are interleukin 17–producing precursors to RORC+CD127+ natural killer–like cells. Nat. Immunol. 10, 66–74 (2008).
Zheng, Y. et al. Interleukin-22 mediates early host defense against attaching and effacing bacterial pathogens. Nat. Med. 14, 282–289 (2008).
Luci, C. et al. Influence of the transcription factor RORγt on the development of NK46+ cell populations in gut and skin. Nat. Immunol. 10, 75–82 (2009).
Klose, C.S., Hoyler, T., Kiss, E.A., Tanriver, Y. & Diefenbach, A. Transcriptional control of innate lymphocyte fate decisions. Curr. Opin. Immunol. 24, 290–296 (2012).
Joshi, N.S. et al. Inflammation directs memory precursor and short-lived effector CD8+ T cell fates via the graded expression of T-bet transcription factor. Immunity 27, 281–295 (2007).
Gordon, S.M. et al. The transcription factors T-bet and Eomes control key checkpoints of natural killer cell maturation. Immunity 36, 55–67 (2012).
Kallies, A. et al. Plasma cell ontogeny defined by quantitative changes in blimp-1 expression. J. Exp. Med. 200, 967–977 (2004).
Garrett, W.S. et al. Communicable ulcerative colitis induced by T-bet deficiency in the innate immune system. Cell 131, 33–45 (2007).
Cherrier, M., Sawa, S. & Eberl, G. Notch, Id2, and RORγt sequentially orchestrate the fetal development of lymphoid tissue inducer cells. J. Exp. Med. 209, 729–740 (2012).
Jackson, J.T. et al. Id2 expression delineates differential checkpoints in the genetic program of CD8α+ and CD103+ dendritic cell lineages. EMBO J. 30, 2690–2704 (2011).
Satoh-Takayama, N. et al. Microbial flora drives interleukin 22 production in intestinal NKp46+ cells that provide innate mucosal immune defense. Immunity 29, 958–970 (2008).
Basu, R. et al. Th22 cells are an important source of IL-22 for host protection against enteropathogenic bacteria. Immunity 37, 1061–1075 (2012).
Vallance, B.A., Deng, W., Knodler, L.A. & Finlay, B.B. Mice lacking T and B lymphocytes develop transient colitis and crypt hyperplasia yet suffer impaired bacterial clearance during Citrobacter rodentium infection. Infect. Immun. 70, 2070–2081 (2002).
Vonarbourg, C. et al. Regulated expression of nuclear receptor RORγt confers distinct functional fates to NK cell receptor-expressing RORγt+ innate lymphocytes. Immunity 33, 736–751 (2010).
Possot, C. et al. Notch signaling is necessary for adult, but not fetal, development of RORγt+ innate lymphoid cells. Nat. Immunol. 12, 949–958 (2011).
Lee, J.S. et al. AHR drives the development of gut ILC22 cells and postnatal lymphoid tissues via pathways dependent on and independent of Notch. Nat. Immunol. 13, 144–151 (2011).
Wei, G. et al. Global mapping of H3K4me3 and H3K27me3 reveals specificity and plasticity in lineage fate determination of differentiating CD4+ T cells. Immunity 30, 155–167 (2009).
Kiss, E.A. et al. Natural aryl hydrocarbon receptor ligands control organogenesis of intestinal lymphoid follicles. Science 334, 1561–1565 (2011).
Qiu, J. et al. The aryl hydrocarbon receptor regulates gut immunity through modulation of innate lymphoid cells. Immunity 36, 92–104 (2012).
Sonnenberg, G.F., Monticelli, L.A., Elloso, M.M., Fouser, L.A. & Artis, D. CD4+ lymphoid tissue-inducer cells promote innate immunity in the gut. Immunity 34, 122–134 (2011).
Lazarevic, V. & Glimcher, L.H. T-bet in disease. Nat. Immunol. 12, 597–606 (2011).
Mombaerts, P. et al. RAG-1-deficient mice have no mature B and T lymphocytes. Cell 68, 869–877 (1992).
Colucci, F. et al. Dissecting NK cell development using a novel alymphoid mouse model: investigating the role of the c-abl proto-oncogene in murine NK cell differentiation. J. Immunol. 162, 2761–2765 (1999).
Finotto, S. et al. Development of spontaneous airway changes consistent with human asthma in mice lacking T-bet. Science 295, 336–338 (2002).
Simon, P. Q-Gene: processing quantitative real-time RT-PCR data. Bioinformatics 19, 1439–1440 (2003).
Nakayamada, S. et al. Early Th1 cell differentiation is marked by a Tfh cell-like transition. Immunity 35, 919–931 (2011).
Acknowledgements
We thank R. Robins-Browne, M. Camilleri, R. Cole, J. Cruickshank and the staff of the animal and flow cytometry facilities of the Walter and Eliza Hall Institute and Medical Research Council Laboratory of Molecular Biology for technical assistance. Supported by the National Health and Medical Research Council of Australia (G.T.B., S.L.N., S.C., L.R. and J.R.G.), the Sylvia and Charles Viertel Foundation (G.T.B.), the Howard Hughes Medical Institute (G.T.B.), the Australian Research Council (S.L.N.), the Leukaemia Foundation (M.J.H.), the American Asthma Foundation (J.A.W.), the Medical Research Council, the American Asthma Foundation (A.N.J.M.) and Victorian State Government Operational Infrastructure Support and Australian Government National Health and Medical Research Council Independent Research Institutes Infrastructure Support Scheme.
Author information
Authors and Affiliations
Contributions
L.C.R., J.R.G., S.C., M.J.H., M.C., S.L.N. and G.T.B. designed and did experiments and analyzed data; L.A.M. did PCR analysis; J.A.W. and A.N.J.M. analyzed nuocytes; and G.T.B., L.C.R., J.R.G. and S.L.N. wrote the paper.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–6 and Tables 1–3 (PDF 1749 kb)
Rights and permissions
About this article
Cite this article
Rankin, L., Groom, J., Chopin, M. et al. The transcription factor T-bet is essential for the development of NKp46+ innate lymphocytes via the Notch pathway. Nat Immunol 14, 389–395 (2013). https://doi.org/10.1038/ni.2545
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ni.2545
This article is cited by
-
The Notch signaling pathway: a potential target for cancer immunotherapy
Journal of Hematology & Oncology (2023)
-
RIPK3 controls MAIT cell accumulation during development but not during infection
Cell Death & Disease (2023)
-
Interferon regulatory factor 1 (IRF-1) promotes intestinal group 3 innate lymphoid responses during Citrobacter rodentium infection
Nature Communications (2022)
-
Regulation of intestinal immunity by dietary fatty acids
Mucosal Immunology (2022)
-
Transcriptional control of mature ILC3 function and plasticity: not just RORγt
Cellular & Molecular Immunology (2022)