Abstract
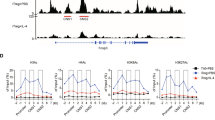
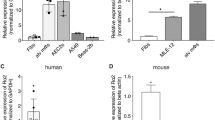
Sox4 is a transcription factor that regulates various developmental processes. Here we show that Sox4 was induced by TGF-β and negatively regulated the transcription factor GATA-3, the master regulator of function of T helper type 2 (TH2) cells, by two distinct mechanisms. First, Sox4 bound directly to GATA-3, preventing its binding to GATA-3 consensus DNA sequences. Second, Sox4 bound to the promoter region of the gene encoding interleukin 5 (IL-5), a TH2 cytokine, and prevented binding of GATA-3 to this promoter. TH2 cell–driven airway inflammation was modulated by alterations in Sox4 expression. Thus, Sox4 acted as a downstream target of TGF-β to inhibit GATA-3 function, TH2 differentiation and TH2 cell–mediated inflammation.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout







Similar content being viewed by others
References
O'Shea, J.J. & Paul, W.E. Mechanisms underlying lineage commitment and plasticity of helper CD4+ T cells. Science 327, 1098–1102 (2010).
Reiner, S.L. Development in motion: helper T cells at work. Cell 129, 33–36 (2007).
Zhu, J., Yamane, H. & Paul, W.E. Differentiation of effector CD4 T cell populations. Annu. Rev. Immunol. 28, 445–489 (2010).
Sakaguchi, S., Yamaguchi, T., Nomura, T. & Ono, M. Regulatory T cells and immune tolerance. Cell 133, 775–787 (2008).
Soroosh, P. & Doherty, T.A. Th9 and allergic disease. Immunology 127, 450–458 (2009).
Witte, E., Witte, K., Warszawska, K., Sabat, R. & Wolk, K. Interleukin-22: A cytokine produced by T, NK and NKT cell subsets, with importance in the innate immune defense and tissue protection. Cytokine Growth Factor Rev. 21, 365–379 (2010).
Korn, T., Bettelli, E., Oukka, M. & Kuchroo, V.K. IL-17 and Th17 cells. Annu. Rev. Immunol. 27, 485–517 (2009).
Dong, C. TH17 cells in development: an updated view of their molecular identity and genetic programming. Nat. Rev. Immunol. 8, 337–348 (2008).
Ho, I.C., Tai, T.S. & Pai, S.Y. GATA3 and the T-cell lineage: essential functions before and after T-helper-2-cell differentiation. Nat. Rev. Immunol. 9, 125–135 (2009).
Zheng, W. & Flavell, R.A. The transcription factor GATA-3 is necessary and sufficient for Th2 cytokine gene expression in CD4 T cells. Cell 89, 587–596 (1997).
Ivanov, I.I. et al. The orphan nuclear receptor RORαt directs the differentiation program of proinflammatory IL-17+ T helper cells. Cell 126, 1121–1133 (2006).
Yang, X.O. et al. T helper 17 lineage differentiation is programmed by orphan nuclear receptors RORα and RORγ. Immunity 28, 29–39 (2008).
Onodera, A. et al. STAT6-mediated displacement of polycomb by trithorax complex establishes long-term maintenance of GATA3 expression in T helper type 2 cells. J. Exp. Med. 207, 2493–2506 (2010).
Wei, L. et al. Discrete roles of STAT4 and STAT6 transcription factors in tuning epigenetic modifications and transcription during T helper cell differentiation. Immunity 32, 840–851 (2010).
Ansel, K.M., Djuretic, I., Tanasa, B. & Rao, A. Regulation of Th2 differentiation and Il4 locus accessibility. Annu. Rev. Immunol. 24, 607–656 (2006).
Wilson, C.B., Rowell, E. & Sekimata, M. Epigenetic control of T-helper-cell differentiation. Nat. Rev. Immunol. 9, 91–105 (2009).
Nakayama, T. & Yamashita, M. Initiation and maintenance of Th2 cell identity. Curr. Opin. Immunol. 20, 265–271 (2008).
Schwenger, G.T. et al. GATA-3 has dual regulatory functions in human interleukin-5 transcription. J. Biol. Chem. 276, 48502–48509 (2001).
Inami, M. et al. CD28 costimulation controls histone hyperacetylation of the interleukin 5 gene locus in developing th2 cells. J. Biol. Chem. 279, 23123–23133 (2004).
Li, M.O., Wan, Y.Y., Sanjabi, S., Robertson, A.K. & Flavell, R.A. Transforming growth factor-β regulation of immune responses. Annu. Rev. Immunol. 24, 99–146 (2006).
Gorelik, L. & Flavell, R.A. Abrogation of TGFβ signaling in T cells leads to spontaneous T cell differentiation and autoimmune disease. Immunity 12, 171–181 (2000).
Nakao, A. et al. Blockade of transforming growth factor β/Smad signaling in T cells by overexpression of Smad7 enhances antigen-induced airway inflammation and airway reactivity. J. Exp. Med. 192, 151–158 (2000).
Gorelik, L., Fields, P.E. & Flavell, R.A. Cutting edge: TGF-β inhibits Th type 2 development through inhibition of GATA-3 expression. J. Immunol. 165, 4773–4777 (2000).
Heath, V.L., Murphy, E.E., Crain, C., Tomlinson, M.G. & O'Garra, A. TGF-β1 down-regulates Th2 development and results in decreased IL-4-induced STAT6 activation and GATA-3 expression. Eur. J. Immunol. 30, 2639–2649 (2000).
Ikushima, H. & Miyazono, K. TGFβ signalling: a complex web in cancer progression. Nat. Rev. Cancer 10, 415–424 (2010).
Smith, J.M. & Koopman, P.A. The ins and outs of transcriptional control: nucleocytoplasmic shuttling in development and disease. Trends Genet. 20, 4–8 (2004).
Lefebvre, V., Dumitriu, B., Penzo-Mendez, A., Han, Y. & Pallavi, B. Control of cell fate and differentiation by Sry-related high-mobility-group box (Sox) transcription factors. Int. J. Biochem. Cell Biol. 39, 2195–2214 (2007).
Schepers, G.E., Teasdale, R.D. & Koopman, P. Twenty pairs of sox: extent, homology, and nomenclature of the mouse and human sox transcription factor gene families. Dev. Cell 3, 167–170 (2002).
Wilson, M. & Koopman, P. Matching SOX: partner proteins and co-factors of the SOX family of transcriptional regulators. Curr. Opin. Genet. Dev. 12, 441–446 (2002).
Schilham, M.W. et al. Defects in cardiac outflow tract formation and pro-B-lymphocyte expansion in mice lacking Sox-4. Nature 380, 711–714 (1996).
Schilham, M.W., Moerer, P., Cumano, A. & Clevers, H.C. Sox-4 facilitates thymocyte differentiation. Eur. J. Immunol. 27, 1292–1295 (1997).
Li, M.O. & Flavell, R.A. TGF-β: a master of all T cell trades. Cell 134, 392–404 (2008).
Lee, H.J. et al. GATA-3 induces T helper cell type 2 (Th2) cytokine expression and chromatin remodeling in committed Th1 cells. J. Exp. Med. 192, 105–115 (2000).
Yamashita, M. et al. Crucial role of MLL for the maintenance of memory T helper type 2 cell responses. Immunity 24, 611–622 (2006).
Penzo-Méndez, A., Dy, P., Pallavi, B. & Lefebvre, V. Generation of mice harboring a Sox4 conditional null allele. Genesis 45, 776–780 (2007).
Ruebel, K.H. et al. Effects of TGFβ1 on gene expression in the HP75 human pituitary tumor cell line identified by gene expression profiling. Endocrine 33, 62–76 (2008).
Ikushima, H. et al. Autocrine TGF-β signaling maintains tumorigenicity of glioma-initiating cells through Sry-related HMG-box factors. Cell Stem Cell 5, 504–514 (2009).
Miaw, S.C., Choi, A., Yu, E., Kishikawa, H. & Ho, I.C. ROG, repressor of GATA, regulates the expression of cytokine genes. Immunity 12, 323–333 (2000).
Fox, A.H., Kowalski, K., King, G.F., Mackay, J.P. & Crossley, M. Key residues characteristic of GATA N-fingers are recognized by FOG. J. Biol. Chem. 273, 33595–33603 (1998).
Hossain, M.B. et al. Lymphoid enhancer factor interacts with GATA-3 and controls its function in T helper type 2 cells. Immunology 125, 377–386 (2008).
Reményi, A. et al. Crystal structure of a POU/HMG/DNA ternary complex suggests differential assembly of Oct4 and Sox2 on two enhancers. Genes Dev. 17, 2048–2059 (2003).
Wissmüller, S., Kosian, T., Wolf, M., Finzsch, M. & Wegner, M. The high-mobility-group domain of Sox proteins interacts with DNA-binding domains of many transcription factors. Nucleic Acids Res. 34, 1735–1744 (2006).
Murakami, A., Shen, H., Ishida, S. & Dickson, C. SOX7 and GATA-4 are competitive activators of Fgf-3 transcription. J. Biol. Chem. 279, 28564–28573 (2004).
Kimura, M. et al. Regulation of Th2 cell differentiation by mel-18, a mammalian polycomb group gene. Immunity 15, 275–287 (2001).
Gorelik, L., Constant, S. & Flavell, R.A. Mechanism of transforming growth factor β-induced inhibition of T helper type 1 differentiation. J. Exp. Med. 195, 1499–1505 (2002).
Smeltz, R.B., Chen, J. & Shevach, E.M. Transforming growth factor-β1 enhances the interferon-γ-dependent, interleukin-12-independent pathway of T helper 1 cell differentiation. Immunology 114, 484–492 (2005).
Dardalhon, V. et al. IL-4 inhibits TGF-β-induced Foxp3+ T cells and, together with TGF-β, generates IL-9+IL-10+Foxp3− effector T cells. Nat. Immunol. 9, 1347–1355 (2008).
Veldhoen, M. et al. Transforming growth factor-β 'reprograms' the differentiation of T helper 2 cells and promotes an interleukin 9–producing subset. Nat. Immunol. 9, 1341–1346 (2008).
Yamashita, M. et al. Bmi1 regulates memory CD4 T cell survival via repression of the Noxa gene. J. Exp. Med. 205, 1109–1120 (2008).
Sussman, J.J., Saito, T., Shevach, E.M., Germain, R.N. & Ashwell, J.D. Thy-1- and Ly-6-mediated lymphokine production and growth inhibition of a T cell hybridoma require co-expression of the T cell antigen receptor complex. J. Immunol. 140, 2520–2526 (1988).
Acknowledgements
We thank D. Loh (Washington University School of Medicine) for DO11.10 mice; R. Kubo, A. Singer and D. Singer for comments and criticisms in the preparation of this manuscript; and K. Sugaya, H. Asou, S. Norikane, M. Kato and T. Ito for technical assistance. Supported by the Global Center for Education and Research in Immune System Regulation and Treatment, City Area Program (Kazusa/Chiba Area) of the Ministry of Education, Culture, Sports, Science and Technology of Japan, Japan Science and Technology Agency Core Research for Evolutional Science and Technology, Japan Science and Technology Agency Precursory Research for Embryonic Science and Technology, the Ministry of Education, Culture, Sports, Science and Technology of Japan (Grants-in-Aid for Scientific Research on Priority Areas 22021008 and 22021011; Scientific Research (B) 21390147 and 23390075), the Ministry of Health, Labor and Welfare of Japan, the Uehara Memorial Foundation, the Mochida Foundation, the Naito Foundation and the Takeda Science Foundation.
Author information
Authors and Affiliations
Contributions
M.K. and M.Y. designed and did experiments, analyzed data and wrote the manuscript; K.S., S.T., A.O., R.S., S.M., D.T., H.H. and C.I. designed and did experiments and edited the manuscript; V.L. established Sox4 deficient mice; and T.N. conceptualized the research, directed the study and wrote and edited the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–7 (PDF 807 kb)
Rights and permissions
About this article
Cite this article
Kuwahara, M., Yamashita, M., Shinoda, K. et al. The transcription factor Sox4 is a downstream target of signaling by the cytokine TGF-β and suppresses TH2 differentiation. Nat Immunol 13, 778–786 (2012). https://doi.org/10.1038/ni.2362
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ni.2362
This article is cited by
-
Regeneration of invariant natural killer T (iNKT) cells: application of iPSC technology for iNKT cell-targeted tumor immunotherapy
Inflammation and Regeneration (2023)
-
Cytokine storm and targeted therapy in hemophagocytic lymphohistiocytosis
Immunologic Research (2022)
-
Immune Inhibitory Properties and Therapeutic Prospects of Transforming Growth Factor-Beta and Interleukin 10 in Autoimmune Hepatitis
Digestive Diseases and Sciences (2022)
-
Sox12 enhances Fbw7-mediated ubiquitination and degradation of GATA3 in Th2 cells
Cellular & Molecular Immunology (2021)
-
Single-cell profiling defines the prognostic benefit of CD39high tissue resident memory CD8+ T cells in luminal-like breast cancer
Communications Biology (2021)