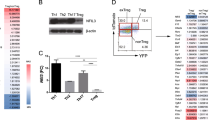
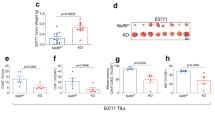
Abstract
Regulatory T cells (Treg cells) are essential for self-tolerance and immune homeostasis. Lack of effector T cell (Teff cell) function and gain of suppressive activity by Treg cells are dependent on the transcriptional program induced by Foxp3. Here we report that repression of SATB1, a genome organizer that regulates chromatin structure and gene expression, was crucial for the phenotype and function of Treg cells. Foxp3, acting as a transcriptional repressor, directly suppressed the SATB1 locus and indirectly suppressed it through the induction of microRNAs that bound the SATB1 3′ untranslated region. Release of SATB1 from the control of Foxp3 in Treg cells caused loss of suppressive function, establishment of transcriptional Teff cell programs and induction of Teff cell cytokines. Our data support the proposal that inhibition of SATB1-mediated modulation of global chromatin remodeling is pivotal for maintaining Treg cell functionality.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout








Similar content being viewed by others
References
Sakaguchi, S., Yamaguchi, T., Nomura, T. & Ono, M. Regulatory T cells and immune tolerance. Cell 133, 775–787 (2008).
Lin, W. et al. Regulatory T cell development in the absence of functional Foxp3. Nat. Immunol. 8, 359–368 (2007).
Wan, Y.Y. & Flavell, R.A. Regulatory T-cell functions are subverted and converted owing to attenuated Foxp3 expression. Nature 445, 766–770 (2007).
Lahl, K. et al. Nonfunctional regulatory T cells and defective control of Th2 cytokine production in natural scurfy mutant mice. J. Immunol. 183, 5662–5672 (2009).
Williams, L.M. & Rudensky, A.Y. Maintenance of the Foxp3-dependent developmental program in mature regulatory T cells requires continued expression of Foxp3. Nat. Immunol. 8, 277–284 (2007).
Fontenot, J.D., Gavin, M.A. & Rudensky, A.Y. Foxp3 programs the development and function of CD4+CD25+ regulatory T cells. Nat. Immunol. 4, 330–336 (2003).
Hori, S., Nomura, T. & Sakaguchi, S. Control of regulatory T cell development by the transcription factor Foxp3. Science 299, 1057–1061 (2003).
O'Shea, J.J. & Paul, W.E. Mechanisms underlying lineage commitment and plasticity of helper CD4v T cells. Science 327, 1098–1102 (2010).
Murphy, K.M. & Stockinger, B. Effector T cell plasticity: flexibility in the face of changing circumstances. Nat. Immunol. 11, 674–680 (2010).
Feuerer, M., Hill, J.A., Mathis, D. & Benoist, C. Foxp3+ regulatory T cells: differentiation, specification, subphenotypes. Nat. Immunol. 10, 689–695 (2009).
Lee, Y.K., Mukasa, R., Hatton, R.D. & Weaver, C.T. Developmental plasticity of Th17 and Treg cells. Curr. Opin. Immunol. 21, 274–280 (2009).
Gavin, M.A. et al. Foxp3-dependent programme of regulatory T-cell differentiation. Nature 445, 771–775 (2007).
Pan, F. et al. Eos mediates Foxp3-dependent gene silencing in CD4+ regulatory T cells. Science 325, 1142–1146 (2009).
Ouyang, W. et al. Foxo proteins cooperatively control the differentiation of Foxp3+ regulatory T cells. Nat. Immunol. 11, 618–627 (2010).
Harada, Y. et al. Transcription factors Foxo3a and Foxo1 couple the E3 ligase Cbl-b to the induction of Foxp3 expression in induced regulatory T cells. J. Exp. Med. 207, 1381–1391 (2010).
Zheng, Y. et al. Regulatory T-cell suppressor program co-opts transcription factor IRF4 to control TH2 responses. Nature 458, 351–356 (2009).
Chaudhry, A. et al. CD4+ regulatory T cells control TH17 responses in a Stat3-dependent manner. Science 326, 986–991 (2009).
Ziegler, S.F. FOXP3: of mice and men. Annu. Rev. Immunol. 24, 209–226 (2006).
Chong, M.M., Rasmussen, J.P., Rudensky, A.Y. & Littman, D.R. The RNAseIII enzyme Drosha is critical in T cells for preventing lethal inflammatory disease. J. Exp. Med. 205, 2005–2017 (2008).
Liston, A., Lu, L.F., O'Carroll, D., Tarakhovsky, A. & Rudensky, A.Y. Dicer-dependent microRNA pathway safeguards regulatory T cell function. J. Exp. Med. 205, 1993–2004 (2008).
Zhou, X. et al. Selective miRNA disruption in T reg cells leads to uncontrolled autoimmunity. J. Exp. Med. 205, 1983–1991 (2008).
Floess, S. et al. Epigenetic control of the foxp3 locus in regulatory T cells. PLoS Biol. 5, e38 (2007).
Li, B. et al. FOXP3 interactions with histone acetyltransferase and class II histone deacetylases are required for repression. Proc. Natl. Acad. Sci. USA 104, 4571–4576 (2007).
Zhou, X., Bailey-Bucktrout, S., Jeker, L.T. & Bluestone, J.A. Plasticity of CD4+ FoxP3+ T cells. Curr. Opin. Immunol. 21, 281–285 (2009).
Alvarez, J.D. et al. The MAR-binding protein SATB1 orchestrates temporal and spatial expression of multiple genes during T-cell development. Genes Dev. 14, 521–535 (2000).
Cai, S., Han, H.J. & Kohwi-Shigematsu, T. Tissue-specific nuclear architecture and gene expression regulated by SATB1. Nat. Genet. 34, 42–51 (2003).
Yasui, D., Miyano, M., Cai, S., Varga-Weisz, P. & Kohwi-Shigematsu, T. SATB1 targets chromatin remodelling to regulate genes over long distances. Nature 419, 641–645 (2002).
Dickinson, L.A., Joh, T., Kohwi, Y. & Kohwi-Shigematsu, T. A tissue-specific MAR/SAR DNA-binding protein with unusual binding site recognition. Cell 70, 631–645 (1992).
Cai, S., Lee, C.C. & Kohwi-Shigematsu, T. SATB1 packages densely looped, transcriptionally active chromatin for coordinated expression of cytokine genes. Nat. Genet. 38, 1278–1288 (2006).
Pfoertner, S. et al. Signatures of human regulatory T cells: an encounter with old friends and new players. Genome Biol. 7, R54 (2006).
Zheng, Y. et al. Genome-wide analysis of Foxp3 target genes in developing and mature regulatory T cells. Nature 445, 936–940 (2007).
Sugimoto, N. et al. Foxp3-dependent and -independent molecules specific for CD25+CD4+ natural regulatory T cells revealed by DNA microarray analysis. Int. Immunol. 18, 1197–1209 (2006).
Lund, R. et al. Identification of genes involved in the initiation of human Th1 or Th2 cell commitment. Eur. J. Immunol. 35, 3307–3319 (2005).
Chen, W. et al. Conversion of peripheral CD4+CD25− naive T cells to CD4+CD25+ regulatory T cells by TGF-beta induction of transcription factor Foxp3. J. Exp. Med. 198, 1875–1886 (2003).
Lahl, K. et al. Selective depletion of Foxp3+ regulatory T cells induces a scurfy-like disease. J. Exp. Med. 204, 57–63 (2007).
Kuczma, M. et al. Foxp3-deficient regulatory T cells do not revert into conventional effector CD4+ T cells but constitute a unique cell subset. J. Immunol. 183, 3731–3741 (2009).
Ishihara, Y., Ito, F. & Shimamoto, N. Increased expression of c-Fos by extracellular signal-regulated kinase activation under sustained oxidative stress elicits BimEL upregulation and hepatocyte apoptosis. FEBS J. 278, 1873–1881 (2011).
Szymczak, A.L. et al. Correction of multi-gene deficiency in vivo using a single 'self-cleaving' 2A peptide-based retroviral vector. Nat. Biotechnol. 22, 589–594 (2004).
Wei, G. et al. Global mapping of H3K4me3 and H3K27me3 reveals specificity and plasticity in lineage fate determination of differentiating CD4+ T cells. Immunity 30, 155–167 (2009).
Lu, L.F. et al. Foxp3-dependent microRNA155 confers competitive fitness to regulatory T cells by targeting SOCS1 protein. Immunity 30, 80–91 (2009).
Kohlhaas, S. et al. Cutting edge: the Foxp3 target miR-155 contributes to the development of regulatory T cells. J. Immunol. 182, 2578–2582 (2009).
Marson, A. et al. Foxp3 occupancy and regulation of key target genes during T-cell stimulation. Nature 445, 931–935 (2007).
Kitoh, A. et al. Indispensable role of the Runx1-Cbfβ transcription complex for in vivo-suppressive function of FoxP3+ regulatory T cells. Immunity 31, 609–620 (2009).
Rudra, D. et al. Runx-CBFβ complexes control expression of the transcription factor Foxp3 in regulatory T cells. Nat. Immunol. 10, 1170–1177 (2009).
Ahlfors, H. et al. SATB1 dictates expression of multiple genes including IL-5 involved in human T helper cell differentiation. Blood 116, 1443–1453 (2010).
Koch, M.A. et al. The transcription factor T-bet controls regulatory T cell homeostasis and function during type 1 inflammation. Nat. Immunol. 10, 595–602 (2009).
Zhou, X. et al. Instability of the transcription factor Foxp3 leads to the generation of pathogenic memory T cells in vivo. Nat. Immunol. 10, 1000–1007 (2009).
Oldenhove, G. et al. Decrease of Foxp3+ Treg cell number and acquisition of effector cell phenotype during lethal infection. Immunity 31, 772–786 (2009).
Brunkow, M.E. et al. Disruption of a new forkhead/winged-helix protein, scurfin, results in the fatal lymphoproliferative disorder of the scurfy mouse. Nat. Genet. 27, 68–73 (2001).
Classen, S. et al. Human resting CD4+ T cells are constitutively inhibited by TGF beta under steady-state conditions. J. Immunol. 178, 6931–6940 (2007).
Sadlon, T.J. et al. Genome-wide identification of human FOXP3 target genes in natural regulatory T cells. J. Immunol. 185, 1071–1081 (2010).
Wiznerowicz, M. & Trono, D. Conditional suppression of cellular genes: lentivirus vector-mediated drug-inducible RNA interference. J. Virol. 77, 8957–8961 (2003).
Tang, Q. et al. In vitro-expanded antigen-specific regulatory T cells suppress autoimmune diabetes. J. Exp. Med. 199, 1455–1465 (2004).
ten Hove, T. et al. Dichotomal role of inhibition of p38 MAPK with SB 203580 in experimental colitis. Gut 50, 507–512 (2002).
Acknowledgements
We thank M. Mai, M. Kraut, S. Keller, N. Kuhn, J. Birke, I. Büchmann, A. Dolf and P. Wurst for technical assistance; M. Hoch, M. Pankratz, S. Burgdorf, A. Popov and A. Staratschek-Jox, as well as all other laboratory members, for discussions; and J. Oldenburg for blood samples from healthy subjects. Supported by the German Research Foundation (Sonderforschungsbereich 832, SFB 704, INST 217/576-1 and INST 217/577-1 to J.L.S. and M.B.), the Wilhelm-Sander-Foundation (J.L.S. and M.B.), the German Cancer Aid (J.L.S.), the German Jose-Carreras-Foundation (J.L.S. and M.B.), the Federal Ministry of Education and Research (Nationale Genomforschungsnetz 2 to J.L.S.), the Humboldt Foundation (J.L.S.), the Leukemia and Lymphoma Society of America (R6029-07 to B.R.B. and K.L.H.), the Juvenile Diabetes Research Foundation (16-2008-643 to X.Z., S.L.B.-B. and J.A.B.), the University of California, San Francisco, Autoimmunity Center of Excellence (X.Z., S.L.B.-B. and J.A.B.), the National Health and Medical Research Council (339123, 565314 to S.C.B.), the German Research Foundation (SCHE 1562 and SFB832 to B.S.), the Juvenile Diabetes Research Foundation Collaborative Centers for Cell Therapy (S.B., T.G. and J.L.R.) and the Juvenile Diabetes Research Foundation Center on Cord Blood Therapies for Type 1 Diabetes (S.B., T.G. and J.L.R.).
Author information
Authors and Affiliations
Contributions
M.B. designed, did and supervised experiments, analyzed data and wrote the manuscript; Y.T. did quantitative PCR, cytometric bead assay, immunoblot analysis, overexpression experiments and filter-retention analysis and analyzed data; R.-U.M. designed and did reporter assays; S.C. did experiments and analyzed data; T.S. did ChIP experiments and analyzed data; K.L. and C.T.M. did experiments with DEREG mice; S.B. and T.G. did overexpression experiments; E.A.S. did and analyzed immunofluorescence experiments; W.K. did histone-methylation studies, S.L.B.-B. and X.Z. did experiments with mice with loxP-flanked Dicer1 alleles; A.H. did bioinformatics analysis; D.S. generated lentivirus contructs; S.D.-P. did microarray experiments; E.E. did flow cytometry sorting; J.B. and A.L. did experiments with Rag2−/− mice; P.A.K. was involved in study design; K.L.H. and B.R.B. provided vital analytical tools; R.B. provided vital analytical tools; T.Q. supervised and analyzed immunofluorescence experiments; C.W. did immunohistochemistry; A.W. did, designed and supervised DNA-methylation experiments; G.M. and M.F. designed and supervised filter-retention experiments; W.K. designed and supervised experiments and wrote the manuscript; B.S. designed and analyzed reporter assays; S.C.B. designed and supervised ChIP experiments; T.S. designed and supervised experiments with DEREG mice and provided vital analytical tools; J.A.B. designed and supervised experiments with mice with loxP-flanked Dicer1 alleles; J.L.R. designed and supervised SATB1-overexpression experiments and wrote the manuscript; J.L.S. designed, supervised and analyzed experiments and wrote the manuscript; and all authors discussed the results and commented on the manuscript.
Corresponding author
Ethics declarations
Competing interests
Research support to J.L.S. and M.B. has been provided in part by Becton Dickinson; R.B. is employed by Becton Dickinson; S.C. is employed by Miltenyi Biotech; and J.L.S., M.B. and R.B. have applied for several US and international patents on Treg cell biology.
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–17, Tables 1–17 and Methods (PDF 2893 kb)
Rights and permissions
About this article
Cite this article
Beyer, M., Thabet, Y., Müller, RU. et al. Repression of the genome organizer SATB1 in regulatory T cells is required for suppressive function and inhibition of effector differentiation. Nat Immunol 12, 898–907 (2011). https://doi.org/10.1038/ni.2084
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ni.2084
This article is cited by
-
Transcriptomic profiling of programmed cell death 1 (PD-1) expressing T cells in early rheumatoid arthritis identifies a decreased CD4 + PD-1 + signature post-treatment
Scientific Reports (2023)
-
The role of transcription factors in shaping regulatory T cell identity
Nature Reviews Immunology (2023)
-
Long Non-Coding RNA ANRIL Regulates Inflammatory Factor Expression in Ulcerative Colitis Via the miR-191-5p/SATB1 Axis
Inflammation (2023)
-
Efficient IL-2R signaling differentially affects the stability, function, and composition of the regulatory T-cell pool
Cellular & Molecular Immunology (2021)
-
Conserved human effector Treg cell transcriptomic and epigenetic signature in arthritic joint inflammation
Nature Communications (2021)