Abstract
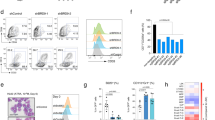
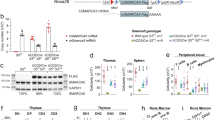
The nuclear adaptor Ldb1 functions as a core component of multiprotein transcription complexes that regulate differentiation in diverse cell types. In the hematopoietic lineage, Ldb1 forms a complex with the non–DNA-binding adaptor Lmo2 and the transcription factors E2A, Scl and GATA-1 (or GATA-2). Here we demonstrate a critical and continuous requirement for Ldb1 in the maintenance of both fetal and adult mouse hematopoietic stem cells (HSCs). Deletion of Ldb1 in hematopoietic progenitors resulted in the downregulation of many transcripts required for HSC maintenance. Genome-wide profiling by chromatin immunoprecipitation followed by sequencing (ChIP-Seq) identified Ldb1 complex–binding sites at highly conserved regions in the promoters of genes involved in HSC maintenance. Our results identify a central role for Ldb1 in regulating the transcriptional program responsible for the maintenance of HSCs.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
Accession codes
References
Boyer, L.A. et al. Core transcriptional regulatory circuitry in human embryonic stem cells. Cell 122, 947–956 (2005).
Loh, Y.H. et al. The Oct4 and Nanog transcription network regulates pluripotency in mouse embryonic stem cells. Nat. Genet. 38, 431–440 (2006).
Akala, O.O. & Clarke, M.F. Hematopoietic stem cell self-renewal. Curr. Opin. Genet. Dev. 16, 496–501 (2006).
Boyer, L.A. et al. Polycomb complexes repress developmental regulators in murine embryonic stem cells. Nature 441, 349–353 (2006).
Majewski, I.J. et al. Polycomb repressive complex 2 (PRC2) restricts hematopoietic stem cell activity. PLoS Biol. 6, e93 (2008).
Matthews, J.M. & Visvader, J.E. LIM-domain-binding protein 1: a multifunctional cofactor that interacts with diverse proteins. EMBO Rep. 4, 1132–1137 (2003).
Wadman, I.A. et al. The LIM-only protein Lmo2 is a bridging molecule assembling an erythroid, DNA-binding complex which includes the TAL1, E47, GATA-1 and Ldb1/NLI proteins. EMBO J. 16, 3145–3157 (1997).
Meier, N. et al. Novel binding partners of Ldb1 are required for haematopoietic development. Development 133, 4913–4923 (2006).
Rodrigues, N.P. et al. Haploinsufficiency of GATA-2 perturbs adult hematopoietic stem-cell homeostasis. Blood 106, 477–484 (2005).
Semerad, C.L., Mercer, E.M., Inlay, M.A., Weissman, I.L. & Murre, C. E2A proteins maintain the hematopoietic stem cell pool and promote the maturation of myelolymphoid and myeloerythroid progenitors. Proc. Natl. Acad. Sci. USA 106, 1930–1935 (2009).
Souroullas, G.P., Salmon, J.M., Sablitzky, F., Curtis, D.J. & Goodell, M.A. Adult hematopoietic stem and progenitor cells require either Lyl1 or Scl for survival. Cell Stem Cell 4, 180–186 (2009).
Warren, A.J. et al. The oncogenic cysteine-rich LIM domain protein rbtn2 is essential for erythroid development. Cell 78, 45–57 (1994).
Ling, K.W. et al. GATA-2 plays two functionally distinct roles during the ontogeny of hematopoietic stem cells. J. Exp. Med. 200, 871–882 (2004).
Tsai, F.Y. et al. An early haematopoietic defect in mice lacking the transcription factor GATA-2. Nature 371, 221–226 (1994).
Mukhopadhyay, M. et al. Functional ablation of the mouse Ldb1 gene results in severe patterning defects during gastrulation. Development 130, 495–505 (2003).
Hwang, M., Gorivodsky, M., Kim, M., Westphal, H. & Geum, D. The neuronal differentiation potential of Ldb1-null mutant embryonic stem cells is dependent on extrinsic influences. Stem Cells 26, 1490–1495 (2008).
Li, L. et al. A requirement for Lim domain binding protein 1 in erythropoiesis. J. Exp. Med. 207, 2543–2550 (2010).
Morrison, S.J., Uchida, N. & Weissman, I.L. The biology of hematopoietic stem cells. Annu. Rev. Cell Dev. Biol. 11, 35–71 (1995).
Lord, B.I. et al. Myeloid cell kinetics in mice treated with recombinant interleukin-3, granulocyte colony-stimulating factor (CSF), or granulocyte-macrophage CSF in vivo. Blood 77, 2154–2159 (1991).
Zhao, Y. et al. LIM-homeodomain proteins Lhx1 and Lhx5, and their cofactor Ldb1, control Purkinje cell differentiation in the developing cerebellum. Proc. Natl. Acad. Sci.USA 104, 13182–13186 (2007).
Kisanuki, Y.Y. et al. Tie2-Cre transgenic mice: a new model for endothelial cell-lineage analysis in vivo. Dev. Biol. 230, 230–242 (2001).
Schlaeger, T.M., Mikkola, H.K., Gekas, C., Helgadottir, H.B. & Orkin, S.H. Tie2Cre-mediated gene ablation defines the stem-cell leukemia gene (SCL/tal1)-dependent window during hematopoietic stem-cell development. Blood 105, 3871–3874 (2005).
Adolfsson, J. et al. Upregulation of Flt3 expression within the bone marrow Lin−Sca1+c-kit+ stem cell compartment is accompanied by loss of self-renewal capacity. Immunity 15, 659–669 (2001).
Yang, L. et al. Identification of Lin−Sca1+ kit+CD34+Flt3− short-term hematopoietic stem cells capable of rapidly reconstituting and rescuing myeloablated transplant recipients. Blood 105, 2717–2723 (2005).
Kiel, M.J. et al. SLAM family receptors distinguish hematopoietic stem and progenitor cells and reveal endothelial niches for stem cells. Cell 121, 1109–1121 (2005).
Kuhn, R., Schwenk, F., Aguet, M. & Rajewsky, K. Inducible gene targeting in mice. Science 269, 1427–1429 (1995).
Wilson, A., Laurenti, E. & Trumpp, A. Balancing dormant and self-renewing hematopoietic stem cells. Curr. Opin. Genet. Dev. 19, 461–468 (2009).
Zon, L.I. Intrinsic and extrinsic control of haematopoietic stem-cell self-renewal. Nature 453, 306–313 (2008).
Lessard, J., Faubert, A. & Sauvageau, G. Genetic programs regulating HSC specification, maintenance and expansion. Oncogene 23, 7199–7209 (2004).
Barski, A. et al. High-resolution profiling of histone methylations in the human genome. Cell 129, 823–837 (2007).
Jothi, R., Cuddapah, S., Barski, A., Cui, K. & Zhao, K. Genome-wide identification of in vivo protein-DNA binding sites from ChIP-Seq data. Nucleic Acids Res. 36, 5221–5231 (2008).
Fujiwara, T. et al. Discovering hematopoietic mechanisms through genome-wide analysis of GATA factor chromatin occupancy. Mol. Cell 36, 667–681 (2009).
Grutz, G.G. et al. The oncogenic T cell LIM-protein Lmo2 forms part of a DNA-binding complex specifically in immature T cells. EMBO J. 17, 4594–4605 (1998).
Hock, H. & Orkin, S.H. Zinc-finger transcription factor Gfi-1: versatile regulator of lymphocytes, neutrophils and hematopoietic stem cells. Curr. Opin. Hematol. 13, 1–6 (2006).
Galan-Caridad, J.M. et al. Zfx controls the self-renewal of embryonic and hematopoietic stem cells. Cell 129, 345–357 (2007).
Kim, I., Saunders, T.L. & Morrison, S.J. Sox17 dependence distinguishes the transcriptional regulation of fetal from adult hematopoietic stem cells. Cell 130, 470–483 (2007).
Anguita, E. et al. Globin gene activation during haemopoiesis is driven by protein complexes nucleated by GATA-1 and GATA-2. EMBO J. 23, 2841–2852 (2004).
Nottingham, W.T. et al. Runx1-mediated hematopoietic stem-cell emergence is controlled by a Gata/Ets/SCL-regulated enhancer. Blood 110, 4188–4197 (2007).
Landry, J.R. et al. Runx genes are direct targets of Scl/Tal1 in the yolk sac and fetal liver. Blood 111, 3005–3014 (2008).
Landry, J.R. et al. Expression of the leukemia oncogene Lmo2 is controlled by an array of tissue-specific elements dispersed over 100 kb and bound by Tal1/Lmo2, Ets, and Gata factors. Blood 113, 5783–5792 (2009).
Wilson, N.K. et al. The transcriptional program controlled by the stem cell leukemia gene Scl/Tal1 during early embryonic hematopoietic development. Blood 113, 5456–5465 (2009).
Gottgens, B. et al. Establishing the transcriptional programme for blood: the SCL stem cell enhancer is regulated by a multiprotein complex containing Ets and GATA factors. EMBO J. 21, 3039–3050 (2002).
Grass, J.A. et al. Distinct functions of dispersed GATA factor complexes at an endogenous gene locus. Mol. Cell. Biol. 26, 7056–7067 (2006).
McCormack, M.P. et al. The lmo2 oncogene initiates leukemia in mice by inducing thymocyte self-renewal. Science 327, 879–883 (2010).
Xu, Z., Huang, S., Chang, L.S., Agulnick, A.D. & Brandt, S.J. Identification of a TAL1 target gene reveals a positive role for the LIM domain-binding protein Ldb1 in erythroid gene expression and differentiation. Mol. Cell. Biol. 23, 7585–7599 (2003).
Acknowledgements
We thank D. El-Khoury and A. Grinberg for technical support, and M. Mukhopadhyay for advice about mouse experiments. Supported by the Intramural Research Program of the National Institutes of Health (Eunice Kennedy Shriver National Institute of Child Health and Human Development, L.L., J.Y.L., T.C., M.G., I.T., Y.Z., H.W. and P.E.L.; National Institute of Environmental Health Sciences, R.J.; National Heart, Lung, and Blood Institute, K.C. and K.Z.) and the National Institutes of Health (DK068634 to E.H.B.).
Author information
Authors and Affiliations
Contributions
L.L. designed and (with assistance from P.E.L.) did all of the experiments; L.L. and P.E.L. designed the study and wrote the manuscript; K.C. and K.Z. assisted in designing and doing the ChIP-Seq experiments; R.J. did the statistical analysis of ChIP-Seq data; E.H.B. provided reagents and input for the ChIP-Seq experiments; J.Y.L., T.C., M.G., I.T., Y.Z. and S.M.H. assisted with specific mouse experiments and provided input on experimental design; and H.W. provided the Ldb1 mouse strains.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–9, Supplementary Tables 1–4 and Supplementary Methods (PDF 8073 kb)
Rights and permissions
About this article
Cite this article
Li, L., Jothi, R., Cui, K. et al. Nuclear adaptor Ldb1 regulates a transcriptional program essential for the maintenance of hematopoietic stem cells. Nat Immunol 12, 129–136 (2011). https://doi.org/10.1038/ni.1978
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ni.1978
This article is cited by
-
LMO2 promotes the development of AML through interaction with transcription co-regulator LDB1
Cell Death & Disease (2023)
-
LMO2 expression is frequent in T-lymphoblastic leukemia and correlates with survival, regardless of T-cell stage
Modern Pathology (2022)
-
3off2: A network reconstruction algorithm based on 2-point and 3-point information statistics
BMC Bioinformatics (2016)
-
BTR: training asynchronous Boolean models using single-cell expression data
BMC Bioinformatics (2016)
-
Neural crest-specific deletion of Ldb1 leads to cleft secondary palate with impaired palatal shelf elevation
BMC Developmental Biology (2014)