Abstract
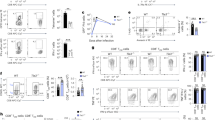
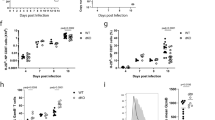
During infection, naive CD8+ T cells differentiate into effector cells, which are armed to eliminate pathogens, and memory cells, which are poised to protect against reinfection. The transcriptional program that regulates terminal differentiation into short-lived effector-memory versus long-lived memory cells is not clearly defined. Through the use of mice expressing reporters for the DNA-binding inhibitors Id2 and Id3, we identified Id3hi precursors of long-lived memory cells before the peak of T cell population expansion or upregulation of cell-surface receptors that indicate memory potential. Deficiency in Id2 or Id3 resulted in loss of distinct CD8+ effector and memory populations, which demonstrated unique roles for these inhibitors of E-protein transcription factors. Furthermore, cytokines altered the expression of Id2 and Id3 differently, which provides insight into how external cues influence gene expression.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout







Similar content being viewed by others
Accession codes
References
Williams, M.A. & Bevan, M.J. Effector and memory CTL differentiation. Annu. Rev. Immunol. 25, 171–192 (2007).
Kaech, S.M., Wherry, E.J. & Ahmed, R. Effector and memory T-cell differentiation: implications for vaccine development. Nat. Rev. Immunol. 2, 251–262 (2002).
Jameson, S.C. & Masopust, D. Diversity in T cell memory: an embarrassment of riches. Immunity 31, 859–871 (2009).
Goldrath, A.W., Luckey, C.J., Park, R., Benoist, C. & Mathis, D. The molecular program induced in T cells undergoing homeostatic proliferation. Proc. Natl. Acad. Sci. USA 101, 16885–16890 (2004).
Kaech, S.M., Hemby, S., Kersh, E. & Ahmed, R. Molecular and functional profiling of memory CD8 T cell differentiation. Cell 111, 837–851 (2002).
Opferman, J.T., Ober, B.T. & Ashton-Rickardt, P.G. Linear differentiation of cytotoxic effectors into memory T lymphocytes. Science 283, 1745–1748 (1999).
Joshi, N.S. et al. Inflammation directs memory precursor and short-lived effector CD8+ T cell fates via the graded expression of T-bet transcription factor. Immunity 27, 281–295 (2007).
Sarkar, S. et al. Functional and genomic profiling of effector CD8 T cell subsets with distinct memory fates. J. Exp. Med. 205, 625–640 (2008).
Ma, A., Koka, R. & Burkett, P. Diverse functions of IL-2, IL-15, and IL-7 in lymphoid homeostasis. Annu. Rev. Immunol. 24, 657–679 (2006).
Badovinac, V.P., Tvinnereim, A.R. & Harty, J.T. Regulation of antigen-specific CD8+ T cell homeostasis by perforin and interferon-gamma. Science 290, 1354–1358 (2000).
Kolumam, G.A., Thomas, S., Thompson, L.J., Sprent, J. & Murali-Krishna, K. Type I interferons act directly on CD8 T cells to allow clonal expansion and memory formation in response to viral infection. J. Exp. Med. 202, 637–650 (2005).
Rubinstein, M.P. et al. IL-7 and IL-15 differentially regulate CD8+ T-cell subsets during contraction of the immune response. Blood 112, 3704–3712 (2008).
Tinoco, R., Alcalde, V., Yang, Y., Sauer, K. & Zuniga, E.I. Cell-intrinsic transforming growth factor-beta signaling mediates virus-specific CD8+ T cell deletion and viral persistence in vivo. Immunity 31, 145–157 (2009).
Badovinac, V.P., Porter, B.B. & Harty, J.T. CD8+ T cell contraction is controlled by early inflammation. Nat. Immunol. 5, 809–817 (2004).
Badovinac, V.P., Messingham, K.A., Jabbari, A., Haring, J.S. & Harty, J.T. Accelerated CD8+ T-cell memory and prime-boost response after dendritic-cell vaccination. Nat. Med. 11, 748–756 (2005).
Wherry, E.J. et al. Lineage relationship and protective immunity of memory CD8 T cell subsets. Nat. Immunol. 4, 225–234 (2003).
Kaech, S.M. et al. Selective expression of the interleukin 7 receptor identifies effector CD8 T cells that give rise to long-lived memory cells. Nat. Immunol. 4, 1191–1198 (2003).
Kalia, V. et al. Prolonged interleukin-2Rα expression on virus-specific CD8+ T cells favors terminal-effector differentiation in vivo. Immunity 32, 91–103 (2010).
Hikono, H. et al. Activation phenotype, rather than central- or effector-memory phenotype, predicts the recall efficacy of memory CD8+ T cells. J. Exp. Med. 204, 1625–1636 (2007).
Hand, T.W., Morre, M. & Kaech, S.M. Expression of IL-7 receptor α is necessary but not sufficient for the formation of memory CD8 T cells during viral infection. Proc. Natl. Acad. Sci. USA 104, 11730–11735 (2007).
Gründemann, C. et al. The NK receptor KLRG1 is dispensable for virus-induced NK and CD8+ T-cell differentiation and function in vivo. Eur. J. Immunol. 40, 1303–1314 (2010).
Williams, M.A., Tyznik, A.J. & Bevan, M.J. Interleukin-2 signals during priming are required for secondary expansion of CD8+ memory T cells. Nature 441, 890–893 (2006).
Pipkin, M.E. et al. Interleukin-2 and inflammation induce distinct transcriptional programs that promote the differentiation of effector cytolytic T cells. Immunity 32, 79–90 (2010).
Mitchell, D.M., Ravkov, E.V. & Williams, M.A. Distinct roles for IL-2 and IL-15 in the differentiation and survival of CD8+ effector and memory T cells. J. Immunol. 184, 6719–6730 (2010).
Kallies, A., Xin, A., Belz, G.T. & Nutt, S.L. Blimp-1 transcription factor is required for the differentiation of effector CD8+ T cells and memory responses. Immunity 31, 283–295 (2009).
Rutishauser, R.L. et al. Transcriptional repressor Blimp-1 promotes CD8+ T cell terminal differentiation and represses the acquisition of central memory T cell properties. Immunity 31, 296–308 (2009).
Sullivan, B.M., Juedes, A., Szabo, S.J., von Herrath, M. & Glimcher, L.H. Antigen-driven effector CD8 T cell function regulated by T-bet. Proc. Natl. Acad. Sci. USA 100, 15818–15823 (2003).
Zhou, X. et al. Differentiation and persistence of memory CD8+ T cells depend on T cell factor 1. Immunity 33, 229–240 (2010).
Banerjee, A. et al. Cutting edge: The transcription factor eomesodermin enables CD8+ T cells to compete for the memory cell niche. J. Immunol. 185, 4988–4992 (2010).
Cannarile, M.A. et al. Transcriptional regulator Id2 mediates CD8+ T cell immunity. Nat. Immunol. 7, 1317–1325 (2006).
Murre, C. Helix-loop-helix proteins and lymphocyte development. Nat. Immunol. 6, 1079–1086 (2005).
Yokota, Y. et al. Development of peripheral lymphoid organs and natural killer cells depends on the helix-loop-helix inhibitor Id2. Nature 397, 702–706 (1999).
Boos, M.D., Yokota, Y., Eberl, G. & Kee, B.L. Mature natural killer cell and lymphoid tissue-inducing cell development requires Id2-mediated suppression of E protein activity. J. Exp. Med. 204, 1119–1130 (2007).
Monticelli, L.A. et al. Transcriptional regulator Id2 controls survival of hepatic NKT cells. Proc. Natl. Acad. Sci. USA 106, 19461–19466 (2009).
Miyazaki, M. et al. The opposing roles of the transcription factor E2A and its antagonist Id3 that orchestrate and enforce the naive fate of T cells. Nat. Immunol. 12, 992–1001 (2011).
Rivera, R.R., Johns, C.P., Quan, J., Johnson, R.S. & Murre, C. Thymocyte selection is regulated by the helix-loop-helix inhibitor protein, Id3. Immunity 12, 17–26 (2000).
Verykokakis, M., Boos, M.D., Bendelac, A. & Kee, B.L. SAP protein-dependent natural killer T-like cells regulate the development of CD8+ T cells with innate lymphocyte characteristics. Immunity 33, 203–215 (2010).
Verykokakis, M. et al. Inhibitor of DNA binding 3 limits development of murine slam-associated adaptor protein-dependent “innate” γδ T cells. PLoS ONE 5, e9303 (2010).
Weinreich, M.A., Odumade, O.A., Jameson, S.C. & Hogquist, K.A. T cells expressing the transcription factor PLZF regulate the development of memory-like CD8+ T cells. Nat. Immunol. 11, 709–716 (2010).
Kee, B.L., Rivera, R.R. & Murre, C. Id3 inhibits B lymphocyte progenitor growth and survival in response to TGF-β. Nat. Immunol. 2, 242–247 (2001).
Martins, G. & Calame, K. Regulation and functions of Blimp-1 in T and B lymphocytes. Annu. Rev. Immunol. 26, 133–169 (2008).
Badovinac, V.P., Haring, J.S. & Harty, J.T. Initial T cell receptor transgenic cell precursor frequency dictates critical aspects of the CD8+ T cell response to infection. Immunity 26, 827–841 (2007).
Lin, Y.C. et al. A global network of transcription factors, involving E2A, EBF1 and Foxo1, that orchestrates B cell fate. Nat. Immunol. 11, 635–643 (2010).
Kedzierska, K. et al. Location rather than CD62L phenotype is critical in the early establishment of influenza-specific CD8+ T cell memory. Proc. Natl. Acad. Sci. USA 104, 9782–9787 (2007).
Schwartz, R., Engel, I., Fallahi-Sichani, M., Petrie, H.T. & Murre, C. Gene expression patterns define novel roles for E47 in cell cycle progression, cytokine-mediated signaling, and T lineage development. Proc. Natl. Acad. Sci. USA 103, 9976–9981 (2006).
D'Cruz, L.M., Knell, J., Fujimoto, J.K. & Goldrath, A.W. An essential role for the transcription factor HEB in thymocyte survival, Tcra rearrangement and the development of natural killer T cells. Nat. Immunol. 11, 240–249 (2010).
Bain, G. et al. Regulation of the helix-loop-helix proteins, E2A and Id3, by the Ras-ERK MAPK cascade. Nat. Immunol. 2, 165–171 (2001).
Savitsky, D., Cimmino, L., Kuo, T., Martins, G.A. & Calame, K. Multiple roles for Blimp-1 in B and T lymphocytes. Adv. Exp. Med. Biol. 596, 9–30 (2007).
Moskowitz, I.P. et al. A molecular pathway including Id2, Tbx5, and Nkx2–5 required for cardiac conduction system development. Cell 129, 1365–1376 (2007).
Agata, Y. et al. Regulation of T cell receptor β gene rearrangements and allelic exclusion by the helix-loop-helix protein, E47. Immunity 27, 871–884 (2007).
Acknowledgements
We thank G. Barton, J. Hamerman, S. Turley, A. Fletcher, M. Rubinstein and members of the Goldrath laboratory for advice and critical review of the manuscript; and L. Shaw for help with figure design. Supported by the Cancer Research Institute, Pew Charitable Trust, the US National Institutes of Health (AI067545 and AI072117 to A.W.G.; and AI073587 to S.S.W.), University of California San Diego (Cellular and Molecular Genetics Training Grant to C.Y.Y.) and the R.E. Bob Smith Education Fund (H.S.L.).
Author information
Authors and Affiliations
Contributions
C.Y.Y. designed and completed the majority of experiments, analyzed and interpreted data, and wrote the manuscript; A.D.S. and R.R.R. generated the reporter mouse lines; J.A.B., J.K., E.Y. and K.C.L. did experiments and discussed and interpreted results; A.K.J. provided technical assistance; H.S.L. and S.S.W. discussed results and provided advice; L.M.D. and C.M. discussed and interpreted results and wrote the manuscript; and A.W.G. directed the study, analyzed and interpreted results, and wrote the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–6 and Table 1 (PDF 1378 kb)
Rights and permissions
About this article
Cite this article
Yang, C., Best, J., Knell, J. et al. The transcriptional regulators Id2 and Id3 control the formation of distinct memory CD8+ T cell subsets. Nat Immunol 12, 1221–1229 (2011). https://doi.org/10.1038/ni.2158
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ni.2158
This article is cited by
-
Id2 epigenetically controls CD8+ T-cell exhaustion by disrupting the assembly of the Tcf3-LSD1 complex
Cellular & Molecular Immunology (2024)
-
Cardinal features of immune memory in innate lymphocytes
Nature Immunology (2023)
-
Regulation of CD8+ T memory and exhaustion by the mTOR signals
Cellular & Molecular Immunology (2023)
-
Hallmarks of CD8+ T cell dysfunction are established within hours of tumor antigen encounter before cell division
Nature Immunology (2023)
-
Chronic lymphocytic leukemia presence impairs antigen-specific CD8+ T-cell responses through epigenetic reprogramming towards short-lived effectors
Leukemia (2023)