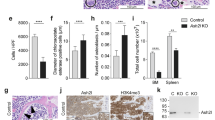
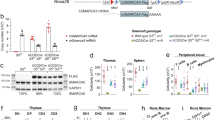
Abstract
Hematopoietic stem cell (HSC) differentiation is regulated by cell-intrinsic and cell-extrinsic cues. In addition to transcriptional regulation, post-translational regulation may also control HSC differentiation. To test this hypothesis, we visualized the ubiquitin-regulated protein stability of a single transcription factor, c-Myc. The stability of c-Myc protein was indicative of HSC quiescence, and c-Myc protein abundance was controlled by the ubiquitin ligase Fbw7. Fine changes in the stability of c-Myc protein regulated the HSC gene-expression signature. Using whole-genome genomic approaches, we identified specific regulators of HSC function directly controlled by c-Myc binding; however, adult HSCs and embryonic stem cells sensed and interpreted c-Myc-regulated gene expression in distinct ways. Our studies show that a ubiquitin ligase–substrate pair can orchestrate the molecular program of HSC differentiation.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout








Similar content being viewed by others
Accession codes
References
Adams, G.B. & Scadden, D.T. The hematopoietic stem cell in its place. Nat. Immunol. 7, 333–337 (2006).
Kiel, M.J. & Morrison, S.J. Uncertainty in the niches that maintain haematopoietic stem cells. Nat. Rev. Immunol. 8, 290–301 (2008).
Kondo, M. et al. Biology of hematopoietic stem cells and progenitors: implications for clinical application. Annu. Rev. Immunol. 21, 759–806 (2003).
Yoshida, T. et al. The role of the chromatin remodeler Mi-2β in hematopoietic stem cell self-renewal and multilineage differentiation. Genes Dev. 22, 1174–1189 (2008).
Moore, K.A. & Lemischka, I.R. Stem cells and their niches. Science 311, 1880–1885 (2006).
Gangaraju, V.K. & Lin, H. MicroRNAs: key regulators of stem cells. Nat. Rev. Mol. Cell Biol. 10, 116–125 (2009).
Cardozo, T. & Pagano, M. The SCF ubiquitin ligase: insights into a molecular machine. Nat. Rev. Mol. Cell Biol. 5, 739–751 (2004).
Harper, J.W. & Schulman, B.A. Structural complexity in ubiquitin recognition. Cell 124, 1133–1136 (2006).
Yamasaki, L. & Pagano, M. Cell cycle, proteolysis and cancer. Curr. Opin. Cell Biol. 16, 623–628 (2004).
Buszczak, M., Paterno, S. & Spradling, A.C. Drosophila stem cells share a common requirement for the histone H2B ubiquitin protease scrawny. Science 323, 248–251 (2009).
Matsuoka, S. et al. Fbxw7 acts as a critical fail-safe against premature loss of hematopoietic stem cells and development of T-ALL. Genes Dev. 22, 986–991 (2008).
Thompson, B.J. et al. Control of hematopoietic stem cell quiescence by the E3 ubiquitin ligase Fbw7. J. Exp. Med. 205, 1395–1408 (2008).
Whetton, A.D. et al. The time is right: proteome biology of stem cells. Cell Stem Cell 2, 215–217 (2008).
Dalla-Favera, R., Martinotti, S., Gallo, R.C., Erikson, J. & Croce, C.M. Translocation and rearrangements of the c-myc oncogene locus in human undifferentiated B-cell lymphomas. Science 219, 963–967 (1983).
O'Neil, J. & Look, A.T. Mechanisms of transcription factor deregulation in lymphoid cell transformation. Oncogene 26, 6838–6849 (2007).
Laurenti, E. et al. Hematopoietic stem cell function and survival depend on c-Myc and N-Myc activity. Cell Stem Cell 3, 611–624 (2008).
Wilson, A. et al. c-Myc controls the balance between hematopoietic stem cell self-renewal and differentiation. Genes Dev. 18, 2747–2763 (2004).
Zhao, X. et al. The HECT-domain ubiquitin ligase Huwe1 controls neural differentiation and proliferation by destabilizing the N-Myc oncoprotein. Nat. Cell Biol. 10, 643–653 (2008).
von der Lehr, N. et al. The F-box protein Skp2 participates in c-Myc proteosomal degradation and acts as a cofactor for c-Myc-regulated transcription. Mol. Cell 11, 1189–1200 (2003).
Welcker, M. et al. The Fbw7 tumor suppressor regulates glycogen synthase kinase 3 phosphorylation-dependent c-Myc protein degradation. Proc. Natl. Acad. Sci. USA 101, 9085–9090 (2004).
Huang, C.Y., Bredemeyer, A.L., Walker, L.M., Bassing, C.H. & Sleckman, B.P. Dynamic regulation of c-Myc proto-oncogene expression during lymphocyte development revealed by a GFP-c-Myc knock-in mouse. Eur. J. Immunol. 38, 342–349 (2008).
Wilson, A. et al. Hematopoietic stem cells reversibly switch from dormancy to self-renewal during homeostasis and repair. Cell 135, 1118–1129 (2008).
Arai, F. et al. Tie2/angiopoietin-1 signaling regulates hematopoietic stem cell quiescence in the bone marrow niche. Cell 118, 149–161 (2004).
Rossi, D.J. et al. Deficiencies in DNA damage repair limit the function of haematopoietic stem cells with age. Nature 447, 725–729 (2007).
Eilers, M. & Eisenman, R.N. Myc's broad reach. Genes Dev. 22, 2755–2766 (2008).
Kuhn, R., Schwenk, F., Aguet, M. & Rajewsky, K. Inducible gene targeting in mice. Science 269, 1427–1429 (1995).
Subramanian, A. et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. USA 102, 15545–15550 (2005).
Georgantas, R.W. III et al. Microarray and serial analysis of gene expression analyses identify known and novel transcripts overexpressed in hematopoietic stem cells. Cancer Res. 64, 4434–4441 (2004).
Ivanova, N.B. et al. A stem cell molecular signature. Science 298, 601–604 (2002).
Goldrath, A.W., Luckey, C.J., Park, R., Benoist, C. & Mathis, D. The molecular program induced in T cells undergoing homeostatic proliferation. Proc. Natl. Acad. Sci. USA 101, 16885–16890 (2004).
Reya, T. et al. A role for Wnt signalling in self-renewal of haematopoietic stem cells. Nature 423, 409–414 (2003).
Karlsson, S. Is TGF-β a stemness regulator? Blood 113, 1208 (2009).
Margolin, A.A. et al. ChIP-on-chip significance analysis reveals large-scale binding and regulation by human transcription factor oncogenes. Proc. Natl. Acad. Sci. USA 106, 244–249 (2009).
Morrison, S.J., Hemmati, H.D., Wandycz, A.M. & Weissman, I.L. The purification and characterization of fetal liver hematopoietic stem cells. Proc. Natl. Acad. Sci. USA 92, 10302–10306 (1995).
Ivanova, N. et al. Dissecting self-renewal in stem cells with RNA interference. Nature 442, 533–538 (2006).
Orford, K.W. & Scadden, D.T. Deconstructing stem cell self-renewal: genetic insights into cell-cycle regulation. Nat. Rev. Genet. 9, 115–128 (2008).
Silva, J. et al. Nanog is the gateway to the pluripotent ground state. Cell 138, 722–737 (2009).
Takahashi, K. et al. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell 131, 861–872 (2007).
Schaniel, C. et al. Delivery of short hairpin RNAs–triggers of gene silencing–into mouse embryonic stem cells. Nat. Methods 3, 397–400 (2006).
Welcker, M. & Clurman, B.E. FBW7 ubiquitin ligase: a tumour suppressor at the crossroads of cell division, growth and differentiation. Nat. Rev. Cancer 8, 83–93 (2008).
Bechard, M. & Dalton, S. Subcellular localization of glycogen synthase kinase 3beta controls embryonic stem cell self-renewal. Mol. Cell. Biol. 29, 2092–2104 (2009).
de Alboran, I.M. et al. Analysis of C-MYC function in normal cells via conditional gene-targeted mutation. Immunity 14, 45–55 (2001).
Demuth, T. et al. MAP-ing glioma invasion: mitogen-activated protein kinase kinase 3 and p38 drive glioma invasion and progression and predict patient survival. Mol. Cancer Ther. 6, 1212–1222 (2007).
Bolstad, B.M., Irizarry, R.A., Astrand, M. & Speed, T.P. A comparison of normalization methods for high density oligonucleotide array data based on variance and bias. Bioinformatics 19, 185–193 (2003).
Saeed, A.I. et al. TM4: a free, open-source system for microarray data management and analysis. Biotechniques 34, 374–378 (2003).
Acknowledgements
We thank the members of the Aifantis Lab and specifically B. King for advice as well as time and effort spent on this manuscript; P. Lopez and the NYU Flow Cytometry Facility for cell sorting; the NYU Cancer Institute Genomics Facility for help with microarray processing; M. Gostissa (Harvard University) and F. Alt (Harvard University) for Mycflox mice; and I. Lemischka and his laboratory (Mount Sinai) for the Nanog-GFP line and technical advice. Supported by the National Institutes of Health (RO1CA133379, RO1CA105129, R21CA141399, R56AI070310 and P30CA016087 to I.A.; RO1AI41428 and RO1AI072039 to B.P.S.; and R01CA120196 to A.F.), the American Cancer Society (RSG0806801 to I.A.), the Edward Mallinckrodt Jr. Foundation, the Irma T. Hirschl Trust, the Alex's Lemonade Stand Foundation (I.A.), the Alexander von Humboldt Foundation (B.A.-O.), the NYU Hematology/Oncology Program (S.M.B.), an NYU Molecular Oncology and Immunology Training Grant (5T32CA009161 to K.C.), the Leukemia & Lymphoma Society (I.A. and A.F.) and the Howard Hughes Medical Institute (I.A.).
Author information
Authors and Affiliations
Contributions
L.R. did most of the experiments and participated in preparing the manuscript; G.D.G., T.P., A.F. and B.A.-O. designed and did the ChIP-plus-microarray and ChIP experiments; K.C., E.L. and B.T. did the ESC experiments; B.P.S., B.T., A.L.B. and B.A.H. generated and did initial studies with the c-Myc–eGFP mice; J.Z. analyzed microarray data; and I.A. designed the study and prepared the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–9 and Supplementary Tables 1–3 (PDF 2270 kb)
Rights and permissions
About this article
Cite this article
Reavie, L., Gatta, G., Crusio, K. et al. Regulation of hematopoietic stem cell differentiation by a single ubiquitin ligase–substrate complex. Nat Immunol 11, 207–215 (2010). https://doi.org/10.1038/ni.1839
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ni.1839
This article is cited by
-
E3 ubiquitin ligase on the biological properties of hematopoietic stem cell
Journal of Molecular Medicine (2023)
-
Ubiquitin ligases: guardians of mammalian development
Nature Reviews Molecular Cell Biology (2022)
-
MYC: a multipurpose oncogene with prognostic and therapeutic implications in blood malignancies
Journal of Hematology & Oncology (2021)
-
Protein quality control of cell stemness
Cell Regeneration (2020)
-
Adaptation to low parasite abundance affects immune investment and immunopathological responses of cavefish
Nature Ecology & Evolution (2020)