Abstract
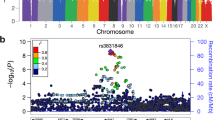
Genome-wide association studies (GWAS) have transformed understanding of susceptibility to testicular germ cell tumors (TGCTs), but much of the heritability remains unexplained. Here we report a new GWAS, a meta-analysis with previous GWAS and a replication series, totaling 7,319 TGCT cases and 23,082 controls. We identify 19 new TGCT risk loci, roughly doubling the number of known TGCT risk loci to 44. By performing in situ Hi-C in TGCT cells, we provide evidence for a network of physical interactions among all 44 TGCT risk SNPs and candidate causal genes. Our findings implicate widespread disruption of developmental transcriptional regulators as a basis of TGCT susceptibility, consistent with failed primordial germ cell differentiation as an initiating step in oncogenesis1. Defective microtubule assembly and dysregulation of KIT–MAPK signaling also feature as recurrently disrupted pathways. Our findings support a polygenic model of risk and provide insight into the biological basis of TGCT.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
References
Manku, G. et al. Changes in the expression profiles of claudins during gonocyte differentiation and in seminomas. Andrology 4, 95–110 (2016).
Le Cornet, C. et al. Testicular cancer incidence to rise by 25% by 2025 in Europe? Model-based predictions in 40 countries using population-based registry data. Eur. J. Cancer 50, 831–839 (2014).
Litchfield, K. et al. Quantifying the heritability of testicular germ cell tumour using both population-based and genomic approaches. Sci. Rep. 5, 13889 (2015).
Swerdlow, A.J., De Stavola, B.L., Swanwick, M.A. & Maconochie, N.E. Risks of breast and testicular cancers in young adult twins in England and Wales: evidence on prenatal and genetic aetiology. Lancet 350, 1723–1728 (1997).
McGlynn, K.A., Devesa, S.S., Graubard, B.I. & Castle, P.E. Increasing incidence of testicular germ cell tumors among black men in the United States. J. Clin. Oncol. 23, 5757–5761 (2005).
Hemminki, K. & Li, X. Familial risk in testicular cancer as a clue to a heritable and environmental aetiology. Br. J. Cancer 90, 1765–1770 (2004).
Kharazmi, E. et al. Cancer risk in relatives of testicular cancer patients by histology type and age at diagnosis: a joint study from five Nordic countries. Eur. Urol. 68, 283–289 (2015).
Rapley, E.A. et al. A genome-wide association study of testicular germ cell tumor. Nat. Genet. 41, 807–810 (2009).
Turnbull, C. & Rahman, N. Genome-wide association studies provide new insights into the genetic basis of testicular germ-cell tumour. Int. J. Androl. 34, e86–e96, discussion e96–e97 (2011).
Kanetsky, P.A. et al. Common variation in KITLG and at 5q31.3 predisposes to testicular germ cell cancer. Nat. Genet. 41, 811–815 (2009).
Turnbull, C. et al. Variants near DMRT1, TERT and ATF7IP are associated with testicular germ cell cancer. Nat. Genet. 42, 604–607 (2010).
Kanetsky, P.A. et al. A second independent locus within DMRT1 is associated with testicular germ cell tumor susceptibility. Hum. Mol. Genet. 20, 3109–3117 (2011).
Ruark, E. et al. Identification of nine new susceptibility loci for testicular cancer, including variants near DAZL and PRDM14. Nat. Genet. 45, 686–689 (2013).
Bojesen, S.E. et al. Multiple independent variants at the TERT locus are associated with telomere length and risks of breast and ovarian cancer. Nat. Genet. 45, 371–384 (2013).
Chung, C.C. et al. Meta-analysis identifies four new loci associated with testicular germ cell tumor. Nat. Genet. 45, 680–685 (2013).
Kristiansen, W. et al. Two new loci and gene sets related to sex determination and cancer progression are associated with susceptibility to testicular germ cell tumor. Hum. Mol. Genet. 24, 4138–4146 (2015).
Litchfield, K. et al. Multi-stage genome-wide association study identifies new susceptibility locus for testicular germ cell tumour on chromosome 3q25. Hum. Mol. Genet. 24, 1169–1176 (2015).
Litchfield, K. et al. Identification of four new susceptibility loci for testicular germ cell tumour. Nat. Commun. 6, 8690 (2015).
Litchfield, K., Shipley, J. & Turnbull, C. Common variants identified in genome-wide association studies of testicular germ cell tumour: an update, biological insights and clinical application. Andrology 3, 34–46 (2015).
Skol, A.D., Scott, L.J., Abecasis, G.R. & Boehnke, M. Joint analysis is more efficient than replication-based analysis for two-stage genome-wide association studies. Nat. Genet. 38, 209–213 (2006).
ENCODE Project Consortium. An integrated encyclopedia of DNA elements in the human genome. Nature 489, 57–74 (2012).
GTEx Consortium. The Genotype-Tissue Expression (GTEx) project. Nat. Genet. 45, 580–585 (2013).
Agnihotri, S. et al. A GATA4-regulated tumor suppressor network represses formation of malignant human astrocytomas. J. Exp. Med. 208, 689–702 (2011).
Hellebrekers, D.M. et al. GATA4 and GATA5 are potential tumor suppressors and biomarkers in colorectal cancer. Clin. Cancer Res. 15, 3990–3997 (2009).
Tsang, A.P. et al. FOG, a multitype zinc finger protein, acts as a cofactor for transcription factor GATA-1 in erythroid and megakaryocytic differentiation. Cell 90, 109–119 (1997).
Adzhubei, I., Jordan, D.M. & Sunyaev, S.R. Predicting functional effect of human missense mutations using PolyPhen-2. Curr. Protoc. Hum. Genet. Chapter 7, Unit 7.20 (2013).
Ketola, I. et al. Developmental expression and spermatogenic stage specificity of transcription factors GATA-1 and GATA-4 and their cofactors FOG-1 and FOG-2 in the mouse testis. Eur. J. Endocrinol. 147, 397–406 (2002).
Zheng, R. & Blobel, G.A. GATA transcription factors and cancer. Genes Cancer 1, 1178–1188 (2010).
Kurimoto, K., Yamaji, M., Seki, Y. & Saitou, M. Specification of the germ cell lineage in mice: a process orchestrated by the PR-domain proteins, Blimp1 and Prdm14. Cell Cycle 7, 3514–3518 (2008).
Ohinata, Y. et al. A signaling principle for the specification of the germ cell lineage in mice. Cell 137, 571–584 (2009).
Yamaji, M. et al. Critical function of Prdm14 for the establishment of the germ cell lineage in mice. Nat. Genet. 40, 1016–1022 (2008).
Smith, C.A., McClive, P.J., Western, P.S., Reed, K.J. & Sinclair, A.H. Conservation of a sex-determining gene. Nature 402, 601–602 (1999).
Rao, S. et al. Differential roles of Sall4 isoforms in embryonic stem cell pluripotency. Mol. Cell. Biol. 30, 5364–5380 (2010).
Greenbaum, M.P. et al. TEX14 is essential for intercellular bridges and fertility in male mice. Proc. Natl. Acad. Sci. USA 103, 4982–4987 (2006).
Mondal, G., Ohashi, A., Yang, L., Rowley, M. & Couch, F.J. Tex14, a Plk1-regulated protein, is required for kinetochore–microtubule attachment and regulation of the spindle assembly checkpoint. Mol. Cell 45, 680–695 (2012).
Jinks, R.N. et al. Recessive nephrocerebellar syndrome on the Galloway–Mowat syndrome spectrum is caused by homozygous protein-truncating mutations of WDR73. Brain 138, 2173–2190 (2015).
Colin, E. et al. Loss-of-function mutations in WDR73 are responsible for microcephaly and steroid-resistant nephrotic syndrome: Galloway–Mowat syndrome. Am. J. Hum. Genet. 95, 637–648 (2014).
Petrovic, A. et al. The MIS12 complex is a protein interaction hub for outer kinetochore assembly. J. Cell Biol. 190, 835–852 (2010).
Rao, C.V., Yamada, H.Y., Yao, Y. & Dai, W. Enhanced genomic instabilities caused by deregulated microtubule dynamics and chromosome segregation: a perspective from genetic studies in mice. Carcinogenesis 30, 1469–1474 (2009).
Barisic, M. et al. Microtubule detyrosination guides chromosomes during mitosis. Science 348, 799–803 (2015).
Ma, W. & Viveiros, M.M. Depletion of pericentrin in mouse oocytes disrupts microtubule organizing center function and meiotic spindle organization. Mol. Reprod. Dev. 81, 1019–1029 (2014).
Litchfield, K. et al. Whole-exome sequencing reveals the mutational spectrum of testicular germ cell tumours. Nat. Commun. 6, 5973 (2015).
Zeron-Medina, J. et al. A polymorphic p53 response element in KIT ligand influences cancer risk and has undergone natural selection. Cell 155, 410–422 (2013).
De Miguel, M.P., Cheng, L., Holland, E.C., Federspiel, M.J. & Donovan, P.J. Dissection of the c-Kit signaling pathway in mouse primordial germ cells by retroviral-mediated gene transfer. Proc. Natl. Acad. Sci. USA 99, 10458–10463 (2002).
Yu, M. et al. The scaffolding adapter Gab2, via Shp-2, regulates kit-evoked mast cell proliferation by activating the Rac/JNK pathway. J. Biol. Chem. 281, 28615–28626 (2006).
Penegar, S. et al. National study of colorectal cancer genetics. Br. J. Cancer 97, 1305–1309 (2007).
Eisen, T., Matakidou, A. & Houlston, R. & GELCAPS Consortium. Identification of low penetrance alleles for lung cancer: the GEnetic Lung CAncer Predisposition Study (GELCAPS). BMC Cancer 8, 244 (2008).
Delaneau, O., Marchini, J. & Zagury, J.F. A linear complexity phasing method for thousands of genomes. Nat. Methods 9, 179–181 (2011).
Howie, B., Fuchsberger, C., Stephens, M., Marchini, J. & Abecasis, G.R. Fast and accurate genotype imputation in genome-wide association studies through pre-phasing. Nat. Genet. 44, 955–959 (2012).
Marchini, J. & Howie, B. Genotype imputation for genome-wide association studies. Nat. Rev. Genet. 11, 499–511 (2010).
Cuppen, E. Genotyping by allele-specific amplification (KASPar). CSH Protoc. 2007, pdb.prot4841 (2007).
Marchini, J., Howie, B., Myers, S., McVean, G. & Donnelly, P. A new multipoint method for genome-wide association studies by imputation of genotypes. Nat. Genet. 39, 906–913 (2007).
Clayton, D.G. et al. Population structure, differential bias and genomic control in a large-scale, case–control association study. Nat. Genet. 37, 1243–1246 (2005).
Liu, J.Z. et al. Meta-analysis and imputation refines the association of 15q25 with smoking quantity. Nat. Genet. 42, 436–440 (2010).
Higgins, J.P. & Thompson, S.G. Quantifying heterogeneity in a meta-analysis. Stat. Med. 21, 1539–1558 (2002).
Scales, M., Jäger, R., Migliorini, G., Houlston, R.S. & Henrion, M.Y. visPIG—a web tool for producing multi-region, multi-track, multi-scale plots of genetic data. PLoS One 9, e107497 (2014).
Pharoah, P.D.P. et al. Polygenic susceptibility to breast cancer and implications for prevention. Nat. Genet. 31, 33–36 (2002).
Cancer Research UK. Testicular cancer statistics. Cancer Research UK http://www.cancerresearchuk.org/health-professional/cancer-statistics/statistics-by-cancer-type/testicular-cancer (accessed 1 May, 2016).
Cowper-Sallari, R. et al. Breast cancer risk-associated SNPs modulate the affinity of chromatin for FOXA1 and alter gene expression. Nat. Genet. 44, 1191–1198 (2012).
Rao, S.S. et al. A 3D map of the human genome at kilobase resolution reveals principles of chromatin looping. Cell 159, 1665–1680 (2014).
Mifsud, B. et al. Mapping long-range promoter contacts in human cells with high-resolution capture Hi-C. Nat. Genet. 47, 598–606 (2015).
Wingett, S. et al. HiCUP: pipeline for mapping and processing Hi-C data. F1000Res. 4, 1310 (2015).
Cairns, J. et al. CHiCAGO: robust detection of DNA looping interactions in Capture Hi-C data. Genome Biol. 17, 127 (2016).
Untergasser, A. et al. Primer3—new capabilities and interfaces. Nucleic Acids Res. 40, e115 (2012).
Schneider, C.A., Rasband, W.S. & Eliceiri, K.W. NIH Image to ImageJ: 25 years of image analysis. Nat. Methods 9, 671–675 (2012).
Ernst, J. & Kellis, M. ChromHMM: automating chromatin-state discovery and characterization. Nat. Methods 9, 215–216 (2012).
Hamosh, A., Scott, A.F., Amberger, J.S., Bocchini, C.A. & McKusick, V.A. Online Mendelian Inheritance in Man (OMIM), a knowledgebase of human genes and genetic disorders. Nucleic Acids Res. 33, D514–D517 (2005).
Ward, L.D. & Kellis, M. HaploReg: a resource for exploring chromatin states, conservation, and regulatory motif alterations within sets of genetically linked variants. Nucleic Acids Res. 40, D930–D934 (2012).
Kheradpour, P. & Kellis, M. Systematic discovery and characterization of regulatory motifs in ENCODE TF binding experiments. Nucleic Acids Res. 42, 2976–2987 (2014).
Zerbino, D.R., Wilder, S.P., Johnson, N., Juettemann, T. & Flicek, P.R. The Ensembl regulatory build. Genome Biol. 16, 56 (2015).
Acknowledgements
We thank the subjects with TGCT and the clinicians involved in their care for participation in this study. We thank the patients and all clinicians forming part of the UK Testicular Cancer Collaboration (UKTCC) for their participation in this study. This study would not have been possible without the contributions of M.K. Bolla (Breast Cancer Association Consortium (BCAC)), Q. Wang (BCAC), K. Michailido (BCAC), J. Dennis (BCAC), P. Hall (Collaborative Oncological Gene-environment Study (COGS)); D.F. Easton (BCAC), A. Berchuck (Ovarian Cancer Association Consortium), R. Eeles (PRACTICAL), G. Chenevix-Trench (The Consortium of Investigators of Modifiers of BRCA1/2), J. Dennis, P. Pharoah, A. Dunning, K. Muir, J. Peto, A. Lee and E. Dicks. We also thank the following for their contributions to this project: J. Simard, P. Kraft, C. Luccarini and the staff of the Centre for Genetic Epidemiology Laboratory; and K.F. Doheny and the staff of the Center for Inherited Disease Research (CIDR) genotyping facility. The results published here are based in part on data generated by the TCGA Research Network. This study makes use of data generated by the Wellcome Trust Case Control Consortium 2 (WTCCC2). We acknowledge the contribution of E. Rapley and M. Stratton to the generation of previously published UK GWAS case data. We thank all the individuals who took part in these studies and all the researchers, clinicians, technicians and administrative staff who have enabled this work to be carried out. We acknowledge National Health Service funding to the National Institute for Health Research Biomedical Research Centre. We thank the UK Genetics of Prostate Cancer Study (UKGPCS) study teams for the recruitment of the UKGPCS controls. Genotyping of the OncoArray was funded by the US National Institutes of Health (NIH) (U19 CA 148537 for Elucidating Loci Involved in Prostate cancer Susceptibility (ELLIPSE) project and X01HG007492 to the Center for Inherited Disease Research (CIDR) under contract number HHSN268201200008I). Additional analytical support was provided by NIH NCI U01 CA188392. The PRACTICAL consortium was supported by Cancer Research UK Grants C5047/A7357, C1287/A10118, C1287/A16563, C5047/A3354, C5047/A10692 and C16913/A6135; the European Commission's Seventh Framework Programme grant agreement 223175 (HEALTH-F2-2009-223175) (D.F.E., R.E. and Z.K.-J.); and the NIH Cancer Post-Cancer GWAS initiative grant 1 U19 CA 148537-01 (the GAME-ON initiative). We thank the following for funding support: the Institute of Cancer Research and the Everyman Campaign, the Prostate Cancer Research Foundation, Prostate Research Campaign UK (now Prostate Action), the Orchid Cancer Appeal, the National Cancer Research Network UK and the National Cancer Research Institute (NCRI) UK. We are grateful for NIHR funding to the Biomedical Research Centre at the Institute of Cancer Research and the Royal Marsden NHS Foundation Trust. We acknowledge funding from the Swedish Cancer Society (CAN2011/484 and CAN2012/823), the Norwegian Cancer Society (grants 418975-71081-PR-2006-0387 and PK01-2007-0375) and the Nordic Cancer Union (grant S-12/07). This study was supported by the Movember Foundation and the Institute of Cancer Research. K.L. is supported by a PhD fellowship from Cancer Research UK. R.S.H. and P.B. are supported by Cancer Research UK (C1298/A8362 Bobby Moore Fund for Cancer Research UK).
Author information
Authors and Affiliations
Consortia
Contributions
C.T., K.L. and R.S.H. designed the study. Case samples were recruited by A.R., R.A.H. and through UKTCC. R.E., A.M.D., K.M., J.P., Z.K.-J., N.P. and D.F.E. supplied OncoArray control data. N.O. administrated genotyping of OncoArray case samples. D.D. coordinated all case sample administration and tracking. K.L., M.L., A.H. and P.B. prepared samples for genotyping experiments. K.L., M.L., G.O., C.L., K.F. and I.A. conducted all Promoter Hi-C and 3C laboratory experiments. C.T., R.S.H. and K.L. designed bioinformatics and statistical analyses. K.L., G.M., C.L. and M.L. conducted all Promoter Hi-C and 3C data analysis. K.L. and P.J.L. conducted transcription factor enrichment analysis. K.L., C.L. and M.L. performed all other bioinformatics and statistical analyses. R.K., T.B.H., W.K., T.G. and F.W. provided Scandinavian GWAS data. K.L. drafted the manuscript with assistance from C.T., R.S.H., M.L., J.S., J.N. and D.T.B. All authors reviewed and contributed to the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Additional information
A list of members appears in the Supplementary Note.
A list of members appears in the Supplementary Note.
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–11, Supplementary Tables 1–10 and Supplementary Note. (PDF 5584 kb)
Rights and permissions
About this article
Cite this article
Litchfield, K., Levy, M., Orlando, G. et al. Identification of 19 new risk loci and potential regulatory mechanisms influencing susceptibility to testicular germ cell tumor. Nat Genet 49, 1133–1140 (2017). https://doi.org/10.1038/ng.3896
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ng.3896
This article is cited by
-
Alternative splicing in lung influences COVID-19 severity and respiratory diseases
Nature Communications (2023)
-
Implementation of individualised polygenic risk score analysis: a test case of a family of four
BMC Medical Genomics (2022)
-
Predicted leukocyte telomere length and risk of germ cell tumours
British Journal of Cancer (2022)
-
Advances in Molecular Profiling and Developing Clinical Trials of CNS Germ Cell Tumors: Present and Future Directions
Current Oncology Reports (2022)
-
A common deletion at BAK1 reduces enhancer activity and confers risk of intracranial germ cell tumors
Nature Communications (2022)