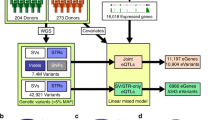
Abstract
The contribution of repetitive elements to quantitative human traits is largely unknown. Here we report a genome-wide survey of the contribution of short tandem repeats (STRs), which constitute one of the most polymorphic and abundant repeat classes, to gene expression in humans. Our survey identified 2,060 significant expression STRs (eSTRs). These eSTRs were replicable in orthogonal populations and expression assays. We used variance partitioning to disentangle the contribution of eSTRs from that of linked SNPs and indels and found that eSTRs contribute 10–15% of the cis heritability mediated by all common variants. Further functional genomic analyses showed that eSTRs are enriched in conserved regions, colocalize with regulatory elements and may modulate certain histone modifications. By analyzing known genome-wide association study (GWAS) signals and searching for new associations in 1,685 whole genomes from deeply phenotyped individuals, we found that eSTRs are enriched in various clinically relevant conditions. These results highlight the contribution of STRs to the genetic architecture of quantitative human traits.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
Accession codes
References
Barrett, J.C. et al. Genome-wide association defines more than 30 distinct susceptibility loci for Crohn's disease. Nat. Genet. 40, 955–962 (2008).
Moffatt, M.F. et al. Genetic variants regulating ORMDL3 expression contribute to the risk of childhood asthma. Nature 448, 470–473 (2007).
GTEx Consortium. The Genotype-Tissue Expression (GTEx) pilot analysis: multitissue gene regulation in humans. Science 348, 648–660 (2015).
Nica, A.C. et al. Candidate causal regulatory effects by integration of expression QTLs with complex trait genetic associations. PLoS Genet. 6, e1000895 (2010).
Nicolae, D.L. et al. Trait-associated SNPs are more likely to be eQTLs: annotation to enhance discovery from GWAS. PLoS Genet. 6, e1000888 (2010).
Ward, L.D. & Kellis, M. Interpreting noncoding genetic variation in complex traits and human disease. Nat. Biotechnol. 30, 1095–1106 (2012).
ENCODE Project Consortium. An integrated encyclopedia of DNA elements in the human genome. Nature 489, 57–74 (2012).
Grundberg, E. et al. Mapping cis- and trans-regulatory effects across multiple tissues in twins. Nat. Genet. 44, 1084–1089 (2012).
Lappalainen, T. et al. Transcriptome and genome sequencing uncovers functional variation in humans. Nature 501, 506–511 (2013).
Stranger, B.E. et al. Relative impact of nucleotide and copy number variation on gene expression phenotypes. Science 315, 848–853 (2007).
Montgomery, S.B. et al. The origin, evolution, and functional impact of short insertion-deletion variants identified in 179 human genomes. Genome Res. 23, 749–761 (2013).
Wright, F.A. et al. Heritability and genomics of gene expression in peripheral blood. Nat. Genet. 46, 430–437 (2014).
Manolio, T.A. et al. Finding the missing heritability of complex diseases. Nature 461, 747–753 (2009).
Press, M.O., Carlson, K.D. & Queitsch, C. The overdue promise of short tandem repeat variation for heritability. Trends Genet. 30, 504–512 (2014).
Ellegren, H. Microsatellites: simple sequences with complex evolution. Nat. Rev. Genet. 5, 435–445 (2004).
Gemayel, R., Vinces, M.D., Legendre, M. & Verstrepen, K.J. Variable tandem repeats accelerate evolution of coding and regulatory sequences. Annu. Rev. Genet. 44, 445–477 (2010).
Weber, J.L. & Wong, C. Mutation of human short tandem repeats. Hum. Mol. Genet. 2, 1123–1128 (1993).
Mirkin, S.M. Expandable DNA repeats and human disease. Nature 447, 932–940 (2007).
Contente, A., Dittmer, A., Koch, M.C., Roth, J. & Dobbelstein, M. A polymorphic microsatellite that mediates induction of PIG3 by p53. Nat. Genet. 30, 315–320 (2002).
Martin, P., Makepeace, K., Hill, S.A., Hood, D.W. & Moxon, E.R. Microsatellite instability regulates transcription factor binding and gene expression. Proc. Natl. Acad. Sci. USA 102, 3800–3804 (2005).
Willems, R., Paul, A., van der Heide, H.G., ter Avest, A.R. & Mooi, F.R. Fimbrial phase variation in Bordetella pertussis: a novel mechanism for transcriptional regulation. EMBO J. 9, 2803–2809 (1990).
Yogev, D., Rosengarten, R., Watson-McKown, R. & Wise, K.S. Molecular basis of Mycoplasma surface antigenic variation: a novel set of divergent genes undergo spontaneous mutation of periodic coding regions and 5′ regulatory sequences. EMBO J. 10, 4069–4079 (1991).
Hefferon, T.W., Groman, J.D., Yurk, C.E. & Cutting, G.R. A variable dinucleotide repeat in the CFTR gene contributes to phenotype diversity by forming RNA secondary structures that alter splicing. Proc. Natl. Acad. Sci. USA 101, 3504–3509 (2004).
Hui, J. et al. Intronic CA-repeat and CA-rich elements: a new class of regulators of mammalian alternative splicing. EMBO J. 24, 1988–1998 (2005).
Rothenburg, S., Koch-Nolte, F., Rich, A. & Haag, F. A polymorphic dinucleotide repeat in the rat nucleolin gene forms Z-DNA and inhibits promoter activity. Proc. Natl. Acad. Sci. USA 98, 8985–8990 (2001).
Weiser, J.N., Love, J.M. & Moxon, E.R. The molecular mechanism of phase variation of H. influenzae lipopolysaccharide. Cell 59, 657–665 (1989).
Vinces, M.D., Legendre, M., Caldara, M., Hagihara, M. & Verstrepen, K.J. Unstable tandem repeats in promoters confer transcriptional evolvability. Science 324, 1213–1216 (2009).
Sureshkumar, S. et al. A genetic defect caused by a triplet repeat expansion in Arabidopsis thaliana. Science 323, 1060–1063 (2009).
Hammock, E.A. & Young, L.J. Microsatellite instability generates diversity in brain and sociobehavioral traits. Science 308, 1630–1634 (2005).
Yáñez-Cuna, J.O. et al. Dissection of thousands of cell type–specific enhancers identifies dinucleotide repeat motifs as general enhancer features. Genome Res. 24, 1147–1156 (2014).
Sawaya, S. et al. Microsatellite tandem repeats are abundant in human promoters and are associated with regulatory elements. PLoS ONE 8, e54710 (2013).
Bilgin Sonay, T. et al. Tandem repeat variation in human and great ape populations and its impact on gene expression divergence. Genome Res. 25, 1591–1599 (2015).
Borel, C. et al. Tandem repeat sequence variation as causative cis-eQTLs for protein-coding gene expression variation: the case of CSTB. Hum. Mutat. 33, 1302–1309 (2012).
Gebhardt, F., Zanker, K.S. & Brandt, B. Modulation of epidermal growth factor receptor gene transcription by a polymorphic dinucleotide repeat in intron 1. J. Biol. Chem. 274, 13176–13180 (1999).
Rockman, M.V. & Wray, G.A. Abundant raw material for cis-regulatory evolution in humans. Mol. Biol. Evol. 19, 1991–2004 (2002).
Shimajiri, S. et al. Shortened microsatellite d(CA)21 sequence down-regulates promoter activity of matrix metalloproteinase 9 gene. FEBS Lett. 455, 70–74 (1999).
Warpeha, K.M. et al. Genotyping and functional analysis of a polymorphic (CCTTT)n repeat of NOS2A in diabetic retinopathy. FASEB J. 13, 1825–1832 (1999).
Hui, J., Stangl, K., Lane, W.S. & Bindereif, A. HnRNP L stimulates splicing of the eNOS gene by binding to variable-length CA repeats. Nat. Struct. Biol. 10, 33–37 (2003).
Sathasivam, K. et al. Aberrant splicing of HTT generates the pathogenic exon 1 protein in Huntington disease. Proc. Natl. Acad. Sci. USA 110, 2366–2370 (2013).
Grünewald, T.G. et al. Chimeric EWSR1-FLI1 regulates the Ewing sarcoma susceptibility gene EGR2 via a GGAA microsatellite. Nat. Genet. 47, 1073–1078 (2015).
1000 Genomes Project Consortium. A map of human genome variation from population-scale sequencing. Nature 467, 1061–1073 (2010).
Willems, T. et al. The landscape of human STR variation. Genome Res. 24, 1894–1904 (2014).
Gymrek, M., Golan, D., Rosset, S. & Erlich, Y. lobSTR: a short tandem repeat profiler for personal genomes. Genome Res. 22, 1154–1162 (2012).
Duyao, M. et al. Trinucleotide repeat length instability and age of onset in Huntington's disease. Nat. Genet. 4, 387–392 (1993).
La Spada, A.R. et al. Meiotic stability and genotype-phenotype correlation of the trinucleotide repeat in X-linked spinal and bulbar muscular atrophy. Nat. Genet. 2, 301–304 (1992).
Flicek, P. et al. Ensembl 2013. Nucleic Acids Res. 41, D48–D55 (2013).
Gebhardt, F., Zanker, K.S. & Brandt, B. Modulation of epidermal growth factor receptor gene transcription by a polymorphic dinucleotide repeat in intron 1. J. Biol. Chem. 274, 13176–13180 (1999).
Stranger, B.E. et al. Patterns of cis regulatory variation in diverse human populations. PLoS Genet. 8, e1002639 (2012).
Payseur, B.A., Place, M. & Weber, J.L. Linkage disequilibrium between STRPs and SNPs across the human genome. Am. J. Hum. Genet. 82, 1039–1050 (2008).
Sawaya, S., Jones, M. & Keller, M. Linkage disequilibrium between single nucleotide polymorphisms and hypermutable loci. bioRxiv 10.1101/020909 (2015).
Lamina, C. et al. A systematic evaluation of short tandem repeats in lipid candidate genes: riding on the SNP-wave. PLoS ONE 9, e102113 (2014).
Gusev, A. et al. Regulatory variants explain much more heritability than coding variants across 11 common diseases. bioRxiv 10.1101/004309 (2014).
Yang, J. et al. Common SNPs explain a large proportion of the heritability for human height. Nat. Genet. 42, 565–569 (2010).
Ioannidis, J.P. Why most discovered true associations are inflated. Epidemiology 19, 640–648 (2008).
Gaffney, D.J. et al. Dissecting the regulatory architecture of gene expression QTLs. Genome Biol. 13, R7 (2012).
Pollard, K.S., Hubisz, M.J., Rosenbloom, K.R. & Siepel, A. Detection of nonneutral substitution rates on mammalian phylogenies. Genome Res. 20, 110–121 (2010).
Trynka, G. et al. Disentangling the effects of colocalizing genomic annotations to functionally prioritize non-coding variants within complex-trait loci. Am. J. Hum. Genet. 97, 139–152 (2015).
Ernst, J. & Kellis, M. ChromHMM: automating chromatin-state discovery and characterization. Nat. Methods 9, 215–216 (2012).
Zeng, H., Hashimoto, T., Kang, D.D. & Gifford, D.K. GERV: a statistical method for generative evaluation of regulatory variants for transcription factor binding. Bioinformatics 10.1093/bioinformatics/btv565 (17 October 2015).
Welter, D. et al. The NHGRI GWAS Catalog, a curated resource of SNP-trait associations. Nucleic Acids Res. 42, D1001–D1006 (2014).
UK10K Consortium. The UK10K project identifies rare variants in health and disease. Nature 526, 82–90 (2015).
Döring, A. et al. SLC2A9 influences uric acid concentrations with pronounced sex-specific effects. Nat. Genet. 40, 430–436 (2008).
Vitart, V. et al. SLC2A9 is a newly identified urate transporter influencing serum urate concentration, urate excretion and gout. Nat. Genet. 40, 437–442 (2008).
Wallace, C. et al. Genome-wide association study identifies genes for biomarkers of cardiovascular disease: serum urate and dyslipidemia. Am. J. Hum. Genet. 82, 139–149 (2008).
Shin, S.-Y. et al. An atlas of genetic influences on human blood metabolites. Nat. Genet. 46, 543–550 (2014).
Weber, J.L. & Broman, K.W. 7 Genotyping for human whole-genome scans: past, present, and future. Adv. Genet. 42, 77–96 (2001).
Chaisson, M.J. et al. Resolving the complexity of the human genome using single-molecule sequencing. Nature 517, 608–611 (2015).
Bhatia, G. et al. Haplotypes of common SNPs can explain missing heritability of complex diseases. bioRxiv 10.1101/022418 (2015).
Purcell, S. et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet. 81, 559–575 (2007).
Guilmatre, A., Highnam, G., Borel, C., Mittelman, D. & Sharp, A.J. Rapid multiplexed genotyping of simple tandem repeats using capture and high-throughput sequencing. Hum. Mutat. 34, 1304–1311 (2013).
Karolchik, D. et al. The UCSC Genome Browser database: 2014 update. Nucleic Acids Res. 42, D764–D770 (2014).
Kent, W.J. et al. The human genome browser at UCSC. Genome Res. 12, 996–1006 (2002).
Trapnell, C., Pachter, L. & Salzberg, S.L. TopHat: discovering splice junctions with RNA-Seq. Bioinformatics 25, 1105–1111 (2009).
Trapnell, C. et al. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat. Protoc. 7, 562–578 (2012).
Stranger, B.E. et al. Population genomics of human gene expression. Nat. Genet. 39, 1217–1224 (2007).
Barbosa-Morais, N.L. et al. A re-annotation pipeline for Illumina BeadArrays: improving the interpretation of gene expression data. Nucleic Acids Res. 38, e17 (2010).
Yang, J., Lee, S.H., Goddard, M.E. & Visscher, P.M. GCTA: a tool for genome-wide complex trait analysis. Am. J. Hum. Genet. 88, 76–82 (2011).
Patterson, N., Price, A.L. & Reich, D. Population structure and eigenanalysis. PLoS Genet. 2, e190 (2006).
Acknowledgements
We thank T. Lappalainen, A. Goren, T. Hashimoto and D. Zielinksi for useful comments and discussions. M.G. was supported by the National Defense Science and Engineering Graduate Fellowship. Y.E. holds a Career Award at the Scientific Interface from the Burroughs Wellcome Fund. This study was supported by a gift from Andria and Paul Heafy (Y.E.), National Institute of Justice (NIJ) grant 2014-DN-BX-K089 (Y.E. and T.W.), and US National Institutes of Health (NIH) grants 1U01HG007037 (H.Z.), R01MH084703 (J.K.P.), R01HG006399 (A.L.P.), HG006696 (A.J.S.), DA033660 (A.J.S.) and MH097018 (A.J.S.) and by research grant 6-FY13-92 from the March of Dimes Foundation (A.J.S.).
Author information
Authors and Affiliations
Contributions
M.G. and Y.E. conceived the study. M.G., T.W., H.Z., B.M. and Y.E. performed analyses. A.G. performed experimental work to generate high-coverage sequencing data for promoter STRs. S.G., M.J.D., A.L.P. and J.K.P. provided statistical input. A.J.S. contributed data and analyses. M.G., T.W. and Y.E. wrote the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Integrated supplementary information
Supplementary Figure 1 STR genotype errors reduce power to detect eSTR associations.
(a) Power to detect associations and (b) estimated variance explained for different simulated values of variance explained by the STR. (black, observed capillary electrophoresis genotypes; blue, lobSTR genotypes).
Supplementary Figure 2 Number of STRs tested per gene.
The histogram gives the number of STRs within 100 kb of each gene that passed quality filters and were included in the eSTR analysis.
Supplementary Figure 3 Unlinked controls follow the null.
QQ plot of association tests between random unlinked STRs and genes.
Supplementary Figure 4 Validation of eSTR analysis using high-coverage genotype calls.
(a) Comparison of STR dosage in low-coverage 1000 Genomes calls versus calls from high-coverage targeted sequencing of promoter STRs. Bubble area represents the number of calls at each data point. For reference, the bubble at (−20, −20) represents 176 calls. “0” denotes the reference allele. The transparent bubble in the center represents calls that are homozygous reference in both data sets. (b) Distribution of the sizes of errors for discordant allele calls. The majority of errors (89.4%) are off by one or two repeat units. (c) Comparison of eSTR effect sizes between the low- and high-coverage data sets. Red dots denote eSTRs with concordant effect directions.
Supplementary Figure 5 Expression values are moderately reproducible across platforms.
(a) Distribution of Spearman rank correlation coefficients between gene expression profiles of individuals measured on microarray versus RNA sequencing platforms. (b) Distribution of Spearman rank correlation coefficients between the order of individuals ranked by expression levels across transcripts measured using microarray versus RNA sequencing platforms.
Supplementary Figure 6 Variance partitioning simulations with a single causal SNP.
Plots show variance partitioning results from simulations in which each gene has a single causal eSNP. (a,b) The distributions of  . Black points denote the true value of the variance explained by the causal SNP. (c,d) The distributions of
. Black points denote the true value of the variance explained by the causal SNP. (c,d) The distributions of  . (a,c) The LMM simulations with STRs as fixed effects. (b,d) The LMM simulations with STRs as random effects. (a–d) Red dots denote the average value of the estimator. Red bars denote the median value of the estimator. The figure shows that the median values of the lead STRs are largely insensitive to the presence of a strong SNP eQTL.
. (a,c) The LMM simulations with STRs as fixed effects. (b,d) The LMM simulations with STRs as random effects. (a–d) Red dots denote the average value of the estimator. Red bars denote the median value of the estimator. The figure shows that the median values of the lead STRs are largely insensitive to the presence of a strong SNP eQTL.
Supplementary Figure 7 Variance partitioning simulations with two causal SNPs.
Plots show variance partitioning results from simulations in which each gene has two causal eSNPs. (a) The distributions of  . Black points denote the true value of the variance explained by the causal SNPs. (b) The distributions of
. Black points denote the true value of the variance explained by the causal SNPs. (b) The distributions of  . Red dots denote the average value of the estimator. Red bars denote the median value of the estimator.
. Red dots denote the average value of the estimator. Red bars denote the median value of the estimator.
Supplementary Figure 8 STR genotype errors cause underestimation of  .
.
The distribution of observed  for each simulated value of
for each simulated value of  is shown for an LMM analysis conducted using true genotypes (black) versus observed genotypes (blue). In the presence of genotyping errors,
is shown for an LMM analysis conducted using true genotypes (black) versus observed genotypes (blue). In the presence of genotyping errors,  is strongly underestimated.
is strongly underestimated.
Supplementary Figure 9 Partitioning variance when treating the STR as a random effect.
The heat map shows the distribution of  and
and  for each gene. Gray lines give the medians of each distribution.
for each gene. Gray lines give the medians of each distribution.
Supplementary Figure 10 Enrichment of eSTRs at promoters and enhancers.
For each distance bin around (a) the TSS and (b) center of H3K27ac peaks, the plot shows the percentage of STRs that were analyzed in that bin that were called as significant eSTRs. (c,d) The number of STRs in each distance bin. Black lines show the number of STRs that were included in our analysis (meaning that they showed sufficient variability and are near genes). Red lines show the number of all STRs in the genome in each bin. Black lines were smoothed by averaging sliding windows of three consecutive data points. In a and b, bins were 10 kb; in c and d, bins were 500 bp.
Supplementary Figure 11 STRs modulate epigenetic signatures.
(a) Schematic of the application of GERV to predict histone modification signatures for different STR alleles. For each eSTR (red) and control STR (gray), we measured the magnitude of the slope between the STR allele and the GERV score and then tested whether the magnitudes were significantly different between the two sets. (b) Comparison of the distribution of slope magnitudes for eSTRs (red) and controls (gray).
Supplementary Figure 12 Enrichment of eSTR genes in GWAS.
Number of eSTR genes (red dashed line) overlapping GWAS genes for each trait. Gray bars give the distribution of the number of overlapping genes from 1,000 control sets of STRs matched on the basis of expression in LCLs and cis heritability. (RA, rheumatoid arthritis; CAD, coronary artery disease; T1D, type 1 diabetes; T2D, type 2 diabetes.)
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–12, Supplementary Note and Supplementary Tables 1–9. (PDF 1826 kb)
Supplementary Data Set 1: Significant eSTRs
A table of all STR × gene associations at a gene-level FDR of 5%. (CSV 18004 kb)
Rights and permissions
About this article
Cite this article
Gymrek, M., Willems, T., Guilmatre, A. et al. Abundant contribution of short tandem repeats to gene expression variation in humans. Nat Genet 48, 22–29 (2016). https://doi.org/10.1038/ng.3461
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ng.3461
This article is cited by
-
Variable number tandem repeats (VNTRs) as modifiers of breast cancer risk in carriers of BRCA1 185delAG
European Journal of Human Genetics (2023)
-
Investigating the relationship between the VNTR variant of the interleukin-1 receptor antagonist gene and coronary in-stent restenosis
Molecular Biology Reports (2023)
-
Comparative genomics reveals the presence of simple sequence repeats in genes related to virulence in plant pathogenic Pythium ultimum and Pythium vexans
Archives of Microbiology (2023)
-
Microsatellite instability assessment is instrumental for Predictive, Preventive and Personalised Medicine: status quo and outlook
EPMA Journal (2023)
-
Genome-wide identification and phenotypic characterization of seizure-associated copy number variations in 741,075 individuals
Nature Communications (2023)