Abstract
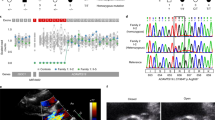
Nonsyndromic mitral valve prolapse (MVP) is a common degenerative cardiac valvulopathy of unknown etiology that predisposes to mitral regurgitation, heart failure and sudden death1. Previous family and pathophysiological studies suggest a complex pattern of inheritance2,3,4,5. We performed a meta-analysis of 2 genome-wide association studies in 1,412 MVP cases and 2,439 controls. We identified 6 loci, which we replicated in 1,422 cases and 6,779 controls, and provide functional evidence for candidate genes. We highlight LMCD1 (LIM and cysteine-rich domains 1), which encodes a transcription factor6 and for which morpholino knockdown of the ortholog in zebrafish resulted in atrioventricular valve regurgitation. A similar zebrafish phenotype was obtained with knockdown of the ortholog of TNS1, which encodes tensin 1, a focal adhesion protein involved in cytoskeleton organization. We also showed expression of tensin 1 during valve morphogenesis and describe enlarged posterior mitral leaflets in Tns1−/− mice. This study identifies the first risk loci for MVP and suggests new mechanisms involved in mitral valve regurgitation, the most common indication for mitral valve repair7.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Freed, L.A. et al. Mitral valve prolapse in the general population: the benign nature of echocardiographic features in the Framingham Heart Study. J. Am. Coll. Cardiol. 40, 1298–1304 (2002).
Disse, S. et al. Mapping of a first locus for autosomal dominant myxomatous mitral valve prolapse to chromosome 16p11.2-p12.1. Am. J. Hum. Genet. 65, 1242–1251 (1999).
Freed, L.A. et al. A locus for autosomal dominant mitral valve prolapse on chromosome 11p15.4. Am. J. Hum. Genet. 72, 1551–1559 (2003).
Kyndt, F. et al. Mapping of X-linked myxomatous valvular dystrophy to chromosome Xq28. Am. J. Hum. Genet. 62, 627–632 (1998).
Nesta, F. et al. New locus for autosomal dominant mitral valve prolapse on chromosome 13: clinical insights from genetic studies. Circulation 112, 2022–2030 (2005).
Rath, N., Wang, Z., Lu, M.M. & Morrisey, E.E. LMCD1/Dyxin is a novel transcriptional cofactor that restricts GATA6 function by inhibiting DNA binding. Mol. Cell. Biol. 25, 8864–8873 (2005).
Avierinos, J.F. et al. Natural history of asymptomatic mitral valve prolapse in the community. Circulation 106, 1355–1361 (2002).
Freed, L.A. et al. Prevalence and clinical outcome of mitral valve prolapse. N. Engl. J. Med. 341, 1–7 (1999).
Devereux, R.B., Brown, W.T., Kramer-Fox, R. & Sachs, I. Inheritance of mitral valve prolapse: effect of age and sex on gene expression. Ann. Intern. Med. 97, 826–832 (1982).
Delling, F.N. et al. Mild expression of mitral valve prolapse in the Framingham offspring: expanding the phenotypic spectrum. J. Am. Soc. Echocardiogr. 27, 17–23 (2014).
Glesby, M.J. & Pyeritz, R.E. Association of mitral valve prolapse and systemic abnormalities of connective tissue. A phenotypic continuum. J. Am. Med. Assoc. 262, 523–528 (1989).
Hagège, A.A. et al. The mitral valve in hypertrophic cardiomyopathy: old versus new concepts. J. Cardiovasc. Transl. Res. 4, 757–766 (2011).
Kyndt, F. et al. Mutations in the gene encoding filamin A as a cause for familial cardiac valvular dystrophy. Circulation 115, 40–49 (2007).
Hall, E.H., Daugherty, A.E., Choi, C.K., Horwitz, A.F. & Brautigan, D.L. Tensin1 requires protein phosphatase-1α in addition to RhoGAP DLC-1 to control cell polarization, migration and invasion. J. Biol. Chem. 284, 34713–34722 (2009).
Sureshbabu, A. et al. IGFBP5 induces cell adhesion, increases cell survival and inhibits cell migration in MCF-7 human breast cancer cells. J. Cell Sci. 125, 1693–1705 (2012).
Chang, C.Y., Lin, S.C., Su, W.H., Ho, C.M. & Jou, Y.S. Somatic LMCD1 mutations promoted cell migration and tumor metastasis in hepatocellular carcinoma. Oncogene 31, 2640–2652 (2012).
Bian, Z.Y. et al. LIM and cysteine-rich domains 1 regulates cardiac hypertrophy by targeting calcineurin–nuclear factor of activated T cells signaling. Hypertension 55, 257–263 (2010).
Beis, D. et al. Genetic and cellular analyses of zebrafish atrioventricular cushion and valve development. Development 132, 4193–4204 (2005).
Schunkert, H. et al. Large-scale association analysis identifies 13 new susceptibility loci for coronary artery disease. Nat. Genet. 43, 333–338 (2011).
Lockhart, M.M. et al. Mef2c regulates transcription of the extracellular matrix protein cartilage link protein 1 in the developing murine heart. PLoS ONE 8, e57073 (2013).
Vasan, R.S. et al. Genetic variants associated with cardiac structure and function: a meta-analysis and replication of genome-wide association data. J. Am. Med. Assoc. 302, 168–178 (2009).
Pyeritz, R.E. & Wappel, M.A. Mitral valve dysfunction in the Marfan syndrome. Clinical and echocardiographic study of prevalence and natural history. Am. J. Med. 74, 797–807 (1983).
Ng, C.M. et al. TGF-β–dependent pathogenesis of mitral valve prolapse in a mouse model of Marfan syndrome. J. Clin. Invest. 114, 1586–1592 (2004).
Rabkin, E. et al. Activated interstitial myofibroblasts express catabolic enzymes and mediate matrix remodeling in myxomatous heart valves. Circulation 104, 2525–2532 (2001).
Lee, A.P. et al. Quantitative analysis of mitral valve morphology in mitral valve prolapse with real-time 3-dimensional echocardiography: importance of annular saddle shape in the pathogenesis of mitral regurgitation. Circulation 127, 832–841 (2013).
Jensen, M.O., Hagege, A.A., Otsuji, Y., Levine, R.A. & Leducq Transatlantic MITRAL Network The unsaddled annulus: biomechanical culprit in mitral valve prolapse? Circulation 127, 766–768 (2013).
Sauls, K. et al. Developmental basis for filamin-A–associated myxomatous mitral valve disease. Cardiovasc. Res. 96, 109–119 (2012).
Levine, R.A. et al. Three-dimensional echocardiographic reconstruction of the mitral valve, with implications for the diagnosis of mitral valve prolapse. Circulation 80, 589–598 (1989).
Levine, R.A., Stathogiannis, E., Newell, J.B., Harrigan, P. & Weyman, A.E. Reconsideration of echocardiographic standards for mitral valve prolapse: lack of association between leaflet displacement isolated to the apical four-chamber view and independent echocardiographic evidence of abnormality. J. Am. Coll. Cardiol. 11, 1010–1019 (1988).
Delaneau, O., Marchini, J. & Zagury, J.F. A linear complexity phasing method for thousands of genomes. Nat. Methods 9, 179–181 (2012).
Marchini, J., Howie, B., Myers, S., McVean, G. & Donnelly, P. A new multipoint method for genome-wide association studies by imputation of genotypes. Nat. Genet. 39, 906–913 (2007).
Li, Y., Willer, C.J., Ding, J., Scheet, P. & Abecasis, G.R. MaCH: using sequence and genotype data to estimate haplotypes and unobserved genotypes. Genet. Epidemiol. 34, 816–834 (2010).
Purcell, S. et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet. 81, 559–575 (2007).
Stouffer, S.A., Suchman, E.A., Devinney, L.C., Star, S.A. & Williams, R.M. The American Soldier: Adjustment During Army Life (Princeton University Press, Princeton, New Jersey, USA, 1949).
Willer, C.J., Li, Y. & Abecasis, G.R. METAL: fast and efficient meta-analysis of genome-wide association scans. Bioinformatics 26, 2190–2191 (2010).
Pruim, R.J. et al. LocusZoom: regional visualization of genome-wide association scan results. Bioinformatics 26, 2336–2337 (2010).
Norris, R.A. et al. Expression of the familial cardiac valvular dystrophy gene, filamin-A, during heart morphogenesis. Dev. Dyn. 239, 2118–2127 (2010).
Milan, D.J., Giokas, A.C., Serluca, F.C., Peterson, R.T. & MacRae, C.A. Notch1b and neuregulin are required for specification of central cardiac conduction tissue. Development 133, 1125–1132 (2006).
Acknowledgements
We acknowledge the major contribution of the Leducq Foundation, Paris for supporting a transatlantic consortium investigating the physiopathology of mitral valve disease, for which this genome-wide association study was a major project (coordinators: R.A.L. and A.A.H.). We thank J. Leyton-Mange, X.-X. Nguyen and M. McLellan for help with the zebrafish experiments and P. Mathieu as one of the main investigators of the PROGRAM (Determinants of the Progression and Outcomes of Organic Mitral Regurgitation) study, from which was ascertained the Canadian MVP case control study. C.D., T.L.T. and J.-J.S. acknowledge a translational research grant on genetics of mitral valve funded by the Nantes Hospital (CHU Nantes). R.A.L. acknowledges grant support from the US National Institutes of Health (NIH; grants K24 HL67434, R01 HL72265 and HL109506). N.B.-N. is recipient of a French young investigator fund (ANR-13-ISV1-0006-0). D.J.M. acknowledges the support of the Hassenfeld Scholar Program and a gift from Michael Zak. P.T.E. is supported by grants from the NIH (HL092577, HL104156, K24HL105780, HL065962), an Established Investigator Award from the American Heart Association (13EIA14220013) and support from the Fondation Leducq (14CVD01). P.P. holds the Canada Research Chair in Valvular Heart Diseases and is supported by grants from the Canadian Institutes of Health Research (CIHR; grants MOP-102737, MOP-114997 and MOP-126072). Y.B. is the recipient of a Junior 2 Research Scholar award from the Fonds de recherche Québec–Santé (FRQS) and is supported by the CIHR (MOP-102481 and MOP-137058). L.F.-F. and J.S. received financial support from the Spanish Society of Cardiology. R.D. was supported by fellowships from the fund for medical discovery of the Massachusetts General Hospital and by a Marie Curie reintegration award from the European Commission R.R.M. and R.A.N. performed their work in a facility constructed with support from the US National Institutes of Health, grant C06 RR018823, from the Extramural Research Facilities Program of the Heart, Lung, and Blood Institute: R01-HL33756 (R.R.M.), COBRE 1P30 GM103342 (R.R.M. and R.A.N.), 8P20 GM103444-07 (R.R.M. and R.A.N.), R01-HL127692 (D.J.M., S.A.S. and R.A.N.) and American Heart Association 15GRNT25080052 (R.A.N.). A.A.H., N.B.-N. and X.J. acknowledge funds from INSERM and the French Society of Cardiology.
Author information
Authors and Affiliations
Consortia
Contributions
C.D., N.B.-N. and X.J. organized and designed the genome-wide association study and the manuscript preparation. A.A.H., R.A.L., S.A.S., H.L.M., P.P., J.S. and L.F.-F. organized and coordinated the network efforts and recruitment of patients. R.D., V.P., T.L.T., F.K., P.P. and Y.B. participated in the recruitment of patients and the interpretation of the echocardiographs. J.-J.S. and X.J. coordinated the collection of patient samples. M.P. and S.L. managed the collection of patient samples and performed DNA extraction and genotyping. C.D. and N.B.-N. conceptualized the statistical analyses. C.D. supervised the statistical analyses. C.D., M.-H.C. and F.S. performed the statistical analyses. D.J.M. conceptualized and supervised the zebrafish experiments. N.T. performed zebrafish experiments, interpreted the data and wrote parts of the manuscript. P.T.E. and E.D. participated in zebrafish experiments and interpretation of data. R.R.M. and R.A.N. conceptualized and supervised the mouse studies and interpreted the data. K.T. performed immunohistochemistry staining of mouse organs. S.H.L. generated the mice. F.N.D., E.J.B., D.Z., M.L., S.H., R. Roussel, F.B., R. Redon, P.F. and R.S.V. conducted the epidemiological studies in control cohorts and/or contributed samples to the GWAS and/or follow-up genotyping. P.B. supervised valve tissues DNA extraction. S.A.S. contributed patient samples and organized follow-up genotyping and interpreted the GWAS and follow-up data. C.D., N.B.-N. and X.J. wrote the manuscript. F.N.D., R.A.N., D.J.M. S.A.S., R.A.L., J.-J.S. and A.A.H. edited the manuscript, and all authors approved its content.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Additional information
A full list of members is provided in the Supplementary Note.
A full list of members is provided in the Supplementary Note.
Integrated supplementary information
Supplementary Figure 2 Igfbp5 expression in mouse developing heart.
Cardiac expression of Igfbp5 (red) was analyzed at embryonic (E13.5), fetal (E17.5) and adult time points. At the time points investigated, very little Igfbp5 protein was detected in the valves. Expression was observed in the myocardium of the left and right atria and the primary atrial septum (PAS) at E13.5 and E17.5 (arrows) as well as the coronary endothelium in the adult (arrow heads). AL, PL, IVS, LV = anterior and posterior mitral leaflets, interventricular septum, and left ventricle, respectively. Green = MF20 (sarcomeric myosin-myocytes), Blue = Hoescht (nuclei).
Supplementary Figure 3 In vivo assessment of mitral valve phenotypes in Tns1 total knockout mice using echocardiographs.
Tensin1+/+, wildtype; Tensin1-/-, Tns1 knockout mice. Images were obtained in a four-chamber view of the heart.
Supplementary Figure 4 In situ hybridizations of tns1 and lmcd1 mRNA distribution in zebrafish embryos.
Top panel shows tns1 expression pattern at 96 hours post fertilization. Expression is found throughout the myocardium, with highest levels in the outflow tract (right). Lower panel shows lmcd1 expression at 72 hours post fertilization (hpf). Expression is found in the entire myocardium, with enhanced expression in the ventricular chamber.
Supplementary Figure 5 In situ hybridizations for developmental markers of valvulogenesis in lmcd1 and tns1 knockdown embryos.
Top panel shows anti-notch1b probe labeling the developing valve in 72-hpf embryos. Lower panel shows anti-bmp4 probe labeling the valve and surrounding myocardium in control (CN) and lmcd1 knockdown embryos. Arrows in both panels point to the approximate location of the AV valve.
Supplementary Figure 6 Pitpnb expression in mouse developing heart.
Cardiac expression of Pitpnb (red) was analyzed at embryonic (E13.5), fetal (E17.5) and adult time points. Pitpnb was detected in valve endothelial and interstitial cells within the mitral leaflets at each of the time points investigated. Expression was also observed in the epicardium (arrow head) and coronary endothelium (arrow) at E17.5. AL, PL, IVS, LV = anterior and posterior mitral leaflets, interventricular septum, and left ventricle, respectively. Green = MF20 (sarcomeric myosin-myocytes), Blue = Hoescht (nuclei).
Supplementary Figure 7 Assessment of cardiac regurgitation in zebrafish morpholino knockdown for genes on Chr17p13.
(a) Genomic context of the association signal observed in the GWAS meta-analysis. (b) Morpholino-mediated knockdown efficacy. Efficacy for smg6 and sgsm2 in embryonic zebrafish was measured by RT-PCR. (c) Fold change in observed mitral regurgitation in 72-hpf zebrafish embryos after morpholino-mediated knockdown. All results are relative to clutchmate controls. n = number of biological replicates per morpholino. (d) Brightfield micrographs displaying gross morphology of 72-hpf embryos following smg6 or sgsm2 knockdown. Scale bar represents 1 mm. CN = control morpholino-injected embryos.
Supplementary Figure 8 Multidimensional scale–based principal-component analysis.
Cases and controls positions on first and second components axis are presented for the two discovery samples (GWAS 1 and GWAS 2). C1, first principal component. C2, second principal component. All individuals plotted are those included in the discovery GWAS who are all French with European ancestry origin. Cases and controls with non-European ancestry based on the principal component analyses were excluded before association tests.
Supplementary Figure 9 Representative gel images from analysis of morpholino efficacy.
rsID indicates the sentinel SNP at the analyzed locus. CN indicates samples amplified from control-injected embryos, whereas MO indicates amplicons from samples obtained following microinjection of gene-specific morpholino. All samples were obtained from 72-hpf embryos.
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–9, Supplementary Tables 1–6 and Supplementary Note. (PDF 2318 kb)
Rights and permissions
About this article
Cite this article
Dina, C., Bouatia-Naji, N., Tucker, N. et al. Genetic association analyses highlight biological pathways underlying mitral valve prolapse. Nat Genet 47, 1206–1211 (2015). https://doi.org/10.1038/ng.3383
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ng.3383
This article is cited by
-
Genome-wide methylation patterns in Marfan syndrome
Clinical Epigenetics (2021)
-
Hyperactivity of Mek in TNS1 knockouts leads to potential treatments for cystic kidney diseases
Cell Death & Disease (2019)
-
Human pre-valvular endocardial cells derived from pluripotent stem cells recapitulate cardiac pathophysiological valvulogenesis
Nature Communications (2019)
-
Follistatin-like 1 in development and human diseases
Cellular and Molecular Life Sciences (2018)
-
Cystinosis (ctns) zebrafish mutant shows pronephric glomerular and tubular dysfunction
Scientific Reports (2017)