Abstract
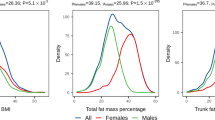
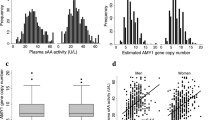
Common multi-allelic copy number variants (CNVs) appear enriched for phenotypic associations compared to their biallelic counterparts1,2,3,4. Here we investigated the influence of gene dosage effects on adiposity through a CNV association study of gene expression levels in adipose tissue. We identified significant association of a multi-allelic CNV encompassing the salivary amylase gene (AMY1) with body mass index (BMI) and obesity, and we replicated this finding in 6,200 subjects. Increased AMY1 copy number was positively associated with both amylase gene expression (P = 2.31 × 10−14) and serum enzyme levels (P < 2.20 × 10−16), whereas reduced AMY1 copy number was associated with increased BMI (change in BMI per estimated copy = −0.15 (0.02) kg/m2; P = 6.93 × 10−10) and obesity risk (odds ratio (OR) per estimated copy = 1.19, 95% confidence interval (CI) = 1.13–1.26; P = 1.46 × 10−10). The OR value of 1.19 per copy of AMY1 translates into about an eightfold difference in risk of obesity between subjects in the top (copy number > 9) and bottom (copy number < 4) 10% of the copy number distribution. Our study provides a first genetic link between carbohydrate metabolism and BMI and demonstrates the power of integrated genomic approaches beyond genome-wide association studies.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
Accession codes
References
Gonzalez, E. et al. The influence of CCL3L1 gene–containing segmental duplications on HIV-1/AIDS susceptibility. Science 307, 1434–1440 (2005).
Hollox, E.J. et al. Psoriasis is associated with increased β-defensin genomic copy number. Nat. Genet. 40, 23–25 (2008).
Fellermann, K. et al. A chromosome 8 gene-cluster polymorphism with low human β-defensin 2 gene copy number predisposes to Crohn disease of the colon. Am. J. Hum. Genet. 79, 439–448 (2006).
Aitman, T.J. et al. Copy number polymorphism in Fcgr3 predisposes to glomerulonephritis in rats and humans. Nature 439, 851–855 (2006).
Eleftherohorinou, H. et al. famCNV: copy number variant association for quantitative traits in families. Bioinformatics 27, 1873–1875 (2011).
Walley, A.J. et al. Differential coexpression analysis of obesity-associated networks in human subcutaneous adipose tissue. Int. J. Obes. (Lond.) 36, 137–147 (2012).
Hysi, P.G. et al. A genome-wide association study for myopia and refractive error identifies a susceptibility locus at 15q25. Nat. Genet. 42, 902–905 (2010).
Field, S.F. et al. Experimental aspects of copy number variant assays at CCL3L1. Nat. Med. 15, 1115–1117 (2009).
Hollox, E.J. in Genomic Structural Variants Vol. 838 (ed. Feuk, L.) 187–207 (Springer New York, 2012).
Aldhous, M.C. et al. Measurement methods and accuracy in copy number variation: failure to replicate associations of β-defensin copy number with Crohn's disease. Hum. Mol. Genet. 19, 4930–4938 (2010).
Carpenter, D., Walker, S., Prescott, N., Schalkwijk, J. & Armour, J.A. Accuracy and differential bias in copy number measurement of CCL3L1 in association studies with three auto-immune disorders. BMC Genomics 12, 418 (2011).
Sudmant, P.H. et al. Diversity of human copy number variation and multicopy genes. Science 330, 641–646 (2010).
Balkau, B., Eschwege, E., Tichet, J. & Marre, M. Proposed criteria for the diagnosis of diabetes: evidence from a French epidemiological study (D.E.S.I.R.). Diabetes Metab. 23, 428–434 (1997).
Spector, T.D. & Williams, F.M. The UK Adult Twin Registry (TwinsUK). Twin Res. Hum. Genet. 9, 899–906 (2006).
Meyre, D. et al. Genome-wide association study for early-onset and morbid adult obesity identifies three new risk loci in European populations. Nat. Genet. 41, 157–159 (2009).
So, H.C., Gui, A.H., Cherny, S.S. & Sham, P.C. Evaluating the heritability explained by known susceptibility variants: a survey of ten complex diseases. Genet. Epidemiol. 35, 310–317 (2011).
Maes, H.H., Neale, M.C. & Eaves, L.J. Genetic and environmental factors in relative body weight and human adiposity. Behav. Genet. 27, 325–351 (1997).
Stunkard, A.J., Foch, T.T. & Hrubec, Z. A twin study of human obesity. J. Am. Med. Assoc. 256, 51–54 (1986).
Wen, W. et al. Meta-analysis identifies common variants associated with body mass index in east Asians. Nat. Genet. 44, 307–311 (2012).
Perry, G.H. et al. Diet and the evolution of human amylase gene copy number variation. Nat. Genet. 39, 1256–1260 (2007).
Chatterton, R.T. Jr., Vogelsong, K.M., Lu, Y.C., Ellman, A.B. & Hudgens, G.A. Salivary α-amylase as a measure of endogenous adrenergic activity. Clin. Physiol. 16, 433–448 (1996).
Park, K.S. Heritability of urine and plasma amylase activity. Jinrui Idengaku Zasshi 22, 79–88 (1977).
Marques-Bonet, T. et al. A burst of segmental duplications in the genome of the African great ape ancestor. Nature 457, 877–881 (2009).
Squires, B.T. Human salivary amylase secretion in relation to diet. J. Physiol. (Lond.) 119, 153–156 (1953).
Mifflin, T.E., Hortin, G. & Bruns, D.E. Electrophoretic assays of amylase isoenzymes and isoforms. Clin. Lab. Med. 6, 583–599 (1986).
Mirski, A. Metabolism of adipose tissue in vitro. Biochem. J. 36, 232–241 (1942).
Bingham, S.A. et al. Nutritional methods in the European Prospective Investigation of Cancer in Norfolk. Public Health Nutr. 4, 847–858 (2001).
Kondo, T., Hayakawa, T., Shibata, T., Sato, Y. & Toda, Y. Serum levels of pancreatic enzymes in lean and obese subjects. Int. J. Pancreatol. 3, 241–248 (1988).
Schneeman, B.O., Inman, M.D. & Stern, J.S. Pancreatic enzyme activity in obese and lean Zucker rats: a developmental study. J. Nutr. 113, 921–925 (1983).
Nakajima, K. et al. Low serum amylase in association with metabolic syndrome and diabetes: a community-based study. Cardiovasc. Diabetol. 10, 34 (2011).
Nakajima, K., Muneyuki, T., Munakata, H. & Kakei, M. Revisiting the cardiometabolic relevance of serum amylase. BMC Res. Notes 4, 419 (2011).
Mandel, A.L. & Breslin, P.A. High endogenous salivary amylase activity is associated with improved glycemic homeostasis following starch ingestion in adults. J. Nutr. 142, 853–858 (2012).
Parks, B.W. et al. Genetic control of obesity and gut microbiota composition in response to high-fat, high-sucrose diet in mice. Cell Metab. 17, 141–152 (2013).
Karlsson, C.L. et al. The microbiota of the gut in preschool children with normal and excessive body weight. Obesity (Silver Spring) 20, 2257–2261 (2012).
Walters, R.G. et al. A new highly penetrant form of obesity due to deletions on chromosome 16p11.2. Nature 463, 671–675 (2010).
Bochukova, E.G. et al. Large, rare chromosomal deletions associated with severe early-onset obesity. Nature 463, 666–670 (2010).
Jacquemont, S. et al. Mirror extreme BMI phenotypes associated with gene dosage at the chromosome 16p11.2 locus. Nature 478, 97–102 (2011).
Willer, C.J. et al. Six new loci associated with body mass index highlight a neuronal influence on body weight regulation. Nat. Genet. 41, 25–34 (2009).
Jarick, I. et al. Novel common copy number variation for early onset extreme obesity on chromosome 11q11 identified by a genome-wide analysis. Hum. Mol. Genet. 20, 840–852 (2011).
Sha, B.Y. et al. Genome-wide association study suggested copy number variation may be associated with body mass index in the Chinese population. J. Hum. Genet. 54, 199–202 (2009).
Speliotes, E.K. et al. Association analyses of 249,796 individuals reveal 18 new loci associated with body mass index. Nat. Genet. 42, 937–948 (2010).
McCarroll, S.A. et al. Integrated detection and population-genetic analysis of SNPs and copy number variation. Nat. Genet. 40, 1166–1174 (2008).
Locke, D.P. et al. Linkage disequilibrium and heritability of copy-number polymorphisms within duplicated regions of the human genome. Am. J. Hum. Genet. 79, 275–290 (2006).
Barnes, C. et al. A robust statistical method for case-control association testing with copy number variation. Nat. Genet. 40, 1245–1252 (2008).
Zhou, B.F. Predictive values of body mass index and waist circumference for risk factors of certain related diseases in Chinese adults—study on optimal cut-off points of body mass index and waist circumference in Chinese adults. Biomed. Environ. Sci. 15, 83–96 (2002).
WHO Expert Consultation. Appropriate body-mass index for Asian populations and its implications for policy and intervention strategies. Lancet 363, 157–163 (2004).
Willer, C.J., Li, Y. & Abecasis, G.R. METAL: fast and efficient meta-analysis of genomewide association scans. Bioinformatics 26, 2190–2191 (2010).
Acknowledgements
The authors would like to thank the study participants in each of the studies, as well as the field investigators for the recruitment and examination of study subjects. The authors would also like to thank P. Maboudou and T. Brousseau for the measurement of serum amylase levels in the ABOS study subjects and P. Gele for the handling of ABOS samples. The authors thank the Genotyping Facility at the Wellcome Trust Sanger Institute for generating the TwinsUK SNP array data. The authors also thank E. Petretto, M. Manca, L. Coin, T. Andrew, I. Prokopenko, T. Norat and S. Richardson for helpful discussions, L. Arnalsteen, H. Verkindt, C. Eberle and M.F. Six for their contributions to the ABOS study and M. Deweider and F. Allegaert for laboratory processing of the DESIR samples, as well as S. Burbidge and M. Harvey of the Imperial College High-Performance Computing service for their assistance (http://www3.imperial.ac.uk/ict/services/hpc).
This work was supported by grants from the Wellcome Trust (grants 079534/z/06/z and 085555), the Medical Research Council (K2010-55X-11285-13), the Swedish Research Council, the Swedish Foundation for Strategic Research to the Sahlgrenska Center for Cardiovascular and Metabolic Research, the Swedish Diabetes Foundation and the Swedish federal government under the LUA/ALF agreement and by the European Community's Seventh Framework Programme (FP7/2007-2013) EUROCHIP project. M.F. is supported by the Medical Research Council (MR/K01353X/1), the British Skin Foundation (5044i), Qatar Foundation (GEQATDIAB) and the Commission of the European Communities (115005). P.F. is supported by the Medical Research Council (G1002084/1), the Imperial College Healthcare National Health Service (NHS) Trust Biomedical Research Centre (P46304 WMDI), the Commission of the European Communities (294785) and the Qatar Foundation (GEQATDIAB).
The DESIR study has been supported by INSERM contracts with CNAMTS, Lilly, Novartis Pharma and Sanofi-Aventis and by INSERM (Réseaux en Santé Publique, Interactions entre les Déterminants de la Santé), Cohortes Santé TGIR, Association Diabète Risque Vasculaire, Fédération Française de Cardiologie, La Fondation de France, ALFEDIAM, ONIVINS, Ardix Medical, Bayer Diagnostics, Becton Dickinson, Cardionics, Merck Santé, Novo Nordisk, Pierre Fabre, Roche and Topcon.
DESIR Study Group. INSERM U780: B.B., M.A. Charles, P. Ducimetière, E. Eschwège; INSERM U367: F. Alhenc-Gelas; CHU (Centre Hospitalier Universitaire) D'Angers: Y. Gallois, A. Girault; Bichat Hospital: F. Fumeron, M. Marre, R. Roussel; CHU Rennes: F. Bonnet; CNRS UMR 8090, Lille: S. Cauchi, P.F.; Centres d'Examens de Santé: Alençon, Angers, Blois, Caen, Chartres, Chateauroux, Cholet, Le Mans, Orleans-Tours; Institute de Recherche Médecine Générale: J. Cogneau; general practitioners of the region; Institute Inter-Regional pour la Santé: C. Born, E. Caces, M. Cailleau, N. Copin, O. Lantieri, J.G. Moreau, F. Rakotozafy, J. Tichet, S. Vol.
This study makes use of data generated by the Genome Structural Variation Consortium (principal investigators N. Carter, M. Hurles, C. Lee and S. Scherer) who we thank for pre-publication access to their CNV discovery (and/or) genotyping data, made available through the websites http://www.sanger.ac.uk/research/areas/humangenetics/cnv/ and http://dgv.tcag.ca/dgv/app/home as a resource to the community. Funding for the project was provided by the Wellcome Trust (grant 077006/Z/05/Z), the Canada Foundation of Innovation and the Ontario Innovation Trust, the Canadian Institutes of Health Research, the Genome Canada/Ontario Genomics Institute, the McLaughlin Centre for Molecular Medicine, the Ontario Ministry of Research and Innovation, the Hospital for Sick Children Foundation, the Department of Pathology at Brigham and Women's Hospital and US National Institutes of Health grants HG004221 and GM081533.
TwinsUK was funded by the Wellcome Trust; the authors acknowledge funding of the GWAS with the support of the Wellcome Trust Sanger Centre and the National Eye Institute via a US National Institutes of Health/Center for Inherited Disease Research (CIDR) genotyping project grant (principal investigator T. Young); also the European Community's Seventh Framework Programme (FP7/2007-2013), ENGAGE project grant agreement HEALTH-F4-2007-201413. The study also receives support from the National Institute for Health Research (NIHR) BioResource Clinical Research Facility and Biomedical Research Centre based at Guy's and St Thomas' NHS Foundation Trust and King's College London. The project also received support from a Biotechnology and Biological Sciences Research Council (BBSRC) project grant (G20234). The ABOS study was partially supported by Lille University Hospital and INSERM (Centre d'Investigation Clinique de Lille) and by grants from Conseil Regional Nord-Pas de Calais (ARCIR Obésité).
F. Pesce was supported by a fellowship from the European Renal Association–European Dialysis and Transplant Association (ERA-EDTA; ALTF 72-2010). M.N.A.-S. is supported by a PhD scholarship from the Research Division of the Qatar Foundation. R.S. is a recipient of a Chercheur Boursier award from the Fonds de la Recherche en Santé du Québec and a New Investigator Award from the Canadian Institutes of Health Research. P.D. is supported by the Wellcome Trust. T.D.S. is in receipt of an NIHR Senior Investigator award and a European Research Council Senior Researcher award.
Author information
Authors and Affiliations
Contributions
M.F. and P.F. conceived the study. M.F., P.F. and T.D.S. directed the project. M.F., J.S.E.-S.M. and P.F. wrote the manuscript. P.T., F. Pesce, A.B. and J.C.A.-A. contributed equally to this work. A.B., T.D.S., R.S., F. Pattou, H.-C.S., P.H.S., L.B., F. Pesce, P.T., R. Dorajoo, P.C.S. and E.E.E. edited the manuscript. J.S.E.-S.M., P.T., F. Pesce, J.C.A.-A., R. Dorajoo, M.N.A.-S., E.O., A.B., A.D. and M.H. performed the laboratory experiments. M.F., J.S.E.-S.M., J.C.A.-A., L.B., P.H.S., E.E.E., P.C.S. and H.-C.S. performed the statistical analyses. R.W.D., A.P., R. Dent, M. Mangino, P.G.H., J.S., M.P., R.C., V.R., E.V., S.F., B.B., M. Marre, S.V.-S., J.W., O.P.-G., P.J., L.S., C.J.H., P.D., R.M., J.L., E.S.T., L.M.S.C., A.W., F. Pattou, T.D.S. and P.F. provided samples, data and/or reagents. All authors commented on and approved the manuscript.
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Text and Figures
Supplementary Tables 1–16, Supplementary Figures 1–17 and Supplementary Note (PDF 3220 kb)
Rights and permissions
About this article
Cite this article
Falchi, M., El-Sayed Moustafa, J., Takousis, P. et al. Low copy number of the salivary amylase gene predisposes to obesity. Nat Genet 46, 492–497 (2014). https://doi.org/10.1038/ng.2939
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ng.2939
This article is cited by
-
A genomics perspective of personalized prevention and management of obesity
Human Genomics (2024)
-
Genetic factors associated with serum amylase in a Japanese population: combined analysis of copy-number and single-nucleotide variants
Journal of Human Genetics (2023)
-
Salivary amylase gene (AMY1) copy number variation has only a minor correlation with body composition in Chinese adults
Genes & Genomics (2023)
-
Impact of salivary and pancreatic amylase gene copy numbers on diabetes, obesity, and functional profiles of microbiome in Northern Japanese population
Scientific Reports (2022)
-
The genetics of obesity: from discovery to biology
Nature Reviews Genetics (2022)