Abstract
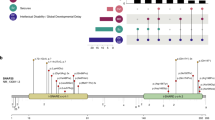
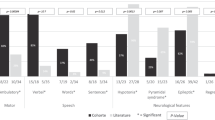
Epilepsy-aphasia syndromes (EAS) are a group of rare, severe epileptic encephalopathies of unknown etiology with a characteristic electroencephalogram (EEG) pattern and developmental regression particularly affecting language. Rare pathogenic deletions that include GRIN2A have been implicated in neurodevelopmental disorders. We sought to delineate the pathogenic role of GRIN2A in 519 probands with epileptic encephalopathies with diverse epilepsy syndromes. We identified four probands with GRIN2A variants that segregated with the disorder in their families. Notably, all four families presented with EAS, accounting for 9% of epilepsy-aphasia cases. We did not detect pathogenic variants in GRIN2A in other epileptic encephalopathies (n = 475) nor in probands with benign childhood epilepsy with centrotemporal spikes (n = 81). We report the first monogenic cause, to our knowledge, for EAS. GRIN2A mutations are restricted to this group of cases, which has important ramifications for diagnostic testing and treatment and provides new insights into the pathogenesis of this debilitating group of conditions.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout


Similar content being viewed by others
Accession codes
References
Berg, A.T. et al. Revised terminology and concepts for organization of seizures and epilepsies: report of the ILAE Commission on Classification and Terminology, 2005–2009. Epilepsia 51, 676–685 (2010).
Tassinari, C.A. et al. Encephalopathy with electrical status epilepticus during slow sleep or ESES syndrome including the acquired aphasia. Clin. Neurophysiol. 111 (suppl. 2), S94–S102 (2000).
Tsai, M.H. et al. Clinical genetic study of the epilepsy-aphasia spectrum. Epilepsia 54, 280–287 (2013).
Deonna, T.W., Roulet, E., Fontan, D. & Marcoz, J.P. Speech and oromotor deficits of epileptic origin in benign partial epilepsy of childhood with rolandic spikes (BPERS). Relationship to the acquired aphasia-epilepsy syndrome. Neuropediatrics 24, 83–87 (1993).
Scheffer, I.E. et al. Autosomal dominant rolandic epilepsy and speech dyspraxia: a new syndrome with anticipation. Ann. Neurol. 38, 633–642 (1995).
Kugler, S.L. et al. An autosomal dominant genetically heterogeneous variant of rolandic epilepsy and speech disorder. Epilepsia 49, 1086–1090 (2008).
Michelucci, R. et al. Familial epilepsy and developmental dysphasia: description of an Italian pedigree with autosomal dominant inheritance and screening of candidate loci. Epilepsy Res. 80, 9–17 (2008).
Roll, P. et al. SRPX2 mutations in disorders of language cortex and cognition. Hum. Mol. Genet. 15, 1195–1207 (2006).
Vears, D.F. et al. Clinical genetic studies in benign childhood epilepsy with centrotemporal spikes. Epilepsia 53, 319–324 (2012).
Lesca, G. et al. Epileptic encephalopathies of the Landau-Kleffner and continuous spike and waves during slow-wave sleep types: genomic dissection makes the link with autism. Epilepsia 53, 1526–1538 (2012).
Reutlinger, C. et al. Deletions in 16p13 including GRIN2A in patients with intellectual disability, various dysmorphic features, and seizure disorders of the rolandic region. Epilepsia 51, 1870–1873 (2010).
Endele, S. et al. Mutations in GRIN2A and GRIN2B encoding regulatory subunits of NMDA receptors cause variable neurodevelopmental phenotypes. Nat. Genet. 42, 1021–1026 (2010).
de Ligt, J. et al. Diagnostic exome sequencing in persons with severe intellectual disability. N. Engl. J. Med. 367, 1921–1929 (2012).
Carvill, G.L. et al. Targeted resequencing in epileptic encephalopathies identifies de novo mutations in CHD2 and SYNGAP1. Nat. Genet. 45, 825–830 (2013).
O'Roak, B.J. et al. Multiplex targeted sequencing identifies recurrently mutated genes in autism spectrum disorders. Science 338, 1619–1622 (2012).
Talukder, I., Borker, P. & Wollmuth, L.P. Specific sites within the ligand-binding domain and ion channel linkers modulate NMDA receptor gating. J. Neurosci. 30, 11792–11804 (2010).
Traynelis, S.F. et al. Glutamate receptor ion channels: structure, regulation, and function. Pharmacol. Rev. 62, 405–496 (2010).
Lesca, G. et al. GRIN2A mutations in acquired epileptic aphasia and related childhood focal epilepsies and encephalopathies with speech and language dysfunction. Nat. Genet. published online; doi:10.1038/ng.2726 (11 August 2013).
O'Roak, B.J. et al. Sporadic autism exomes reveal a highly interconnected protein network of de novo mutations. Nature 485, 246–250 (2012).
Acknowledgements
We thank the subjects and their families for participating in our research. H.C.M. is supported by a grant from the US National Institutes of Health (NIH; NINDS 1R01NS069605) and is a recipient of a Burroughs Wellcome Fund Career Award for Medical Scientists. This work was supported by the National Health and Medical Research Council of Australia (Program Grant 628952 to S.F.B. and I.E.S., Practitioner Fellowship 1006110 to I.E.S. and CJ Martin Fellowship (546493) to M.S.H.) and by a Health Research Council of New Zealand project grant to L.G.S. P.S. is supported by ANR (Agence Nationale de la Recherche) grant EPILAND with EuroBiomed label, and P.S., N. Burnashev and N. Bruneau are supported by INSERM.
Author information
Authors and Affiliations
Contributions
G.L.C., H.C.M. and I.E.S. designed the study and wrote the manuscript. H.C.M. and I.E.S. supervised the study. G.L.C. constructed libraries, developed the variant calling pipeline (assisted by J.C.), analyzed sequence data, conducted RNA transcript analysis (assisted by E.G.) and performed haplotyping. J.C. and G.L.C. performed aCGH. A.K. performed mutation segregation analysis. B.J.O. and J.S. developed the MIP methodology and analysis pipeline. B.M.R., S.C.Y., L.G.S., S.J.T., M.-H.T. and R.W. performed phenotypic analysis. R.O., J.A.D. and M.S.H. conducted mutation screening in the BECTS cohort. B.M.R., S.F.B. and I.E.S. critically reviewed the manuscript. N.L., N. Bruneau, N. Burnashev and P.S. generated mutant transcripts and performed single-channel recordings and analysis.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1 and 2 and Supplementary Tables 1–3 (PDF 1954 kb)
Rights and permissions
About this article
Cite this article
Carvill, G., Regan, B., Yendle, S. et al. GRIN2A mutations cause epilepsy-aphasia spectrum disorders. Nat Genet 45, 1073–1076 (2013). https://doi.org/10.1038/ng.2727
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ng.2727
This article is cited by
-
Rare genetic brain disorders with overlapping neurological and psychiatric phenotypes
Nature Reviews Neurology (2024)
-
Therapeutic potential of N-methyl-D-aspartate receptor modulators in psychiatry
Neuropsychopharmacology (2024)
-
GRIN2A mutation is a novel indicator of stratifying beneficiaries of immune checkpoint inhibitors in multiple cancers
Cancer Gene Therapy (2024)
-
Differential functional consequences of GRIN2A mutations associated with schizophrenia and neurodevelopmental disorders
Scientific Reports (2024)
-
Genetik und genetische Diagnostik fokaler Epilepsien des Kindesalters – Was? Wann? Warum?
Clinical Epileptology (2024)