Abstract
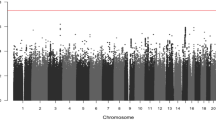
We conducted a genome-wide scan of SNPs to identify variants associated with length of survival in 1,331 individuals with esophageal squamous-cell carcinoma (ESCC), with associations validated in 2 independent sets including 1,962 individuals with this cancer. We identified rs1050631 in SLC39A6 as associated with the survival times of affected individuals, with the hazard ratio for death from ESCC in the combined sample being 1.30 (95% confidence interval (CI) = 1.19−1.43; P = 3.77 × 10−8). rs7242481, located in the 5′ UTR of SLC39A6, disturbs a transcriptional repressor binding site and results in upregulation of SLC39A6 expression. Immunohistochemical staining of ESCC tissues showed that higher expression of SLC39A6 protein was correlated with shorter length of survival in individuals with advanced ESCC (P = 0.013). Knockdown of SLC39A6 expression suppressed proliferation and invasion in ESCC cells. These results suggest that SLC39A6 has an important role in the prognosis of ESCC and may be a potential therapeutic target.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Zhao, P., Dai, M., Chen, W. & Li, N. Cancer trends in China. Jpn. J. Clin. Oncol. 40, 281–285 (2010).
Zhang, D.W. et al. Operable squamous esophageal carcinoma: current results from the East. World J. Surg. 18, 347–354 (1994).
Wang, L.S. et al. Prognosis of esophageal squamous cell carcinoma: analysis of clinicopathological and biological factors. Am. J. Gastroenterol. 94, 1933–1940 (1999).
Liu, J. et al. Which factors are associated with actual 5-year survival of oesophageal squamous cell carcinoma? Eur. J. Cardiothorac. Surg. 41, e7–e11 (2012).
Lee, P.C., Port, J.L., Paul, S., Stiles, B.M. & Altorki, N.K. Predictors of long-term survival after resection of esophageal carcinoma with nonregional nodal metastases. Ann. Thorac. Surg. 88, 186–192 (2009).
Sun, Z.G. & Wang, Z. Clinical study on lymph node metastatic recurrence in patients with N0 esophageal squamous cell cancer. Dis. Esophagus 24, 182–188 (2011).
Okumura, H. et al. Prognostic factors in esophageal squamous cell carcinoma patients treated with neoadjuvant chemoradiation therapy. Int. J. Clin. Oncol. 18, 329–334 (2013).
Stiles, B.M. et al. Predictors of survival in patients with persistent nodal metastases after preoperative chemotherapy for esophageal cancer. J. Thorac. Cardiovasc. Surg. 139, 387–394 (2010).
Thrift, A.P. et al. The influence of prediagnostic demographic and lifestyle factors on esophageal squamous cell carcinoma survival. Int. J. Cancer 130, 2407–2416 (2012).
Azzato, E.M. et al. Association between a germline OCA2 polymorphism at chromosome 15q13.1 and estrogen receptor–negative breast cancer survival. J. Natl. Cancer Inst. 102, 650–662 (2010).
Wu, C. et al. Genome-wide interrogation identifies YAP1 variants associated with survival of small-cell lung cancer patients. Cancer Res. 70, 9721–9729 (2010).
Wu, X. et al. Genome-wide association study of survival in non-small cell lung cancer patients receiving platinum-based chemotherapy. J. Natl. Cancer Inst. 103, 817–825 (2011).
Lu, C., Xie, H., Wang, F., Shen, H. & Wang, J. Diet folate, DNA methylation and genetic polymorphisms of MTHFR C677T in association with the prognosis of esophageal squamous cell carcinoma. BMC Cancer 11, 91 (2011).
Boonstra, J.J., van Marion, R., Tilanus, H.W. & Dinjens, W.N. Functional polymorphisms associated with disease-free survival in resected carcinoma of the esophagus. J. Gastrointest. Surg. 15, 48–56 (2011).
Kuwahara, A. et al. TNFRSF1B A1466G genotype is predictive of clinical efficacy after treatment with a definitive 5-fluorouracil/cisplatin-based chemoradiotherapy in Japanese patients with esophageal squamous cell carcinoma. J. Exp. Clin. Cancer Res. 29, 100 (2010).
Okuno, T. et al. Favorable genetic polymorphisms predictive of clinical outcome of chemoradiotherapy for stage II/III esophageal squamous cell carcinoma in Japanese. Am. J. Clin. Oncol. 30, 252–257 (2007).
Kaneko, K. et al. EGFR gene alterations as a prognostic biomarker in advanced esophageal squamous cell carcinoma. Front. Biosci. 15, 65–72 (2010).
Innocenti, F. et al. A genome-wide association study of overall survival in pancreatic cancer patients treated with gemcitabine in CALGB 80303. Clin. Cancer Res. 18, 577–584 (2012).
Willis, J.A. et al. A replication study and genome-wide scan of single nucleotide polymorphisms associated with pancreatic cancer risk and overall survival. Clin. Cancer Res. 18, 3942–3951 (2012).
Wu, C. et al. Genome-wide association study of survival in patients with pancreatic adenocarcinoma. Gut published online; doi:10.1136/gutjnl-2012-303477 (24 November 2012).
Shu, X.O. et al. Novel genetic markers of breast cancer survival identified by a genome-wide association study. Cancer Res. 72, 1182–1189 (2012).
Cui, R. et al. Functional variants in ADH1B and ALDH2 coupled with alcohol and smoking synergistically enhance esophageal cancer risk. Gastroenterology 137, 1768–1775 (2009).
Wang, L.D. et al. Genome-wide association study of esophageal squamous cell carcinoma in Chinese subjects identifies susceptibility loci at PLCE1 and C20orf54. Nat. Genet. 42, 759–763 (2010).
Abnet, C.C. et al. A shared susceptibility locus in PLCE1 at 10q23 for gastric adenocarcinoma and esophageal squamous cell carcinoma. Nat. Genet. 42, 764–767 (2010).
Wu, C. et al. Genome-wide association study identifies three new susceptibility loci for esophageal squamous-cell carcinoma in Chinese populations. Nat. Genet. 43, 679–684 (2011).
Taylor, K.M. & Nicholson, R.I. The LZT proteins; the LIV-1 subfamily of zinc transporters. Biochim. Biophys. Acta 1611, 16–30 (2003).
John, E. et al. Zinc in innate and adaptive tumor immunity. J. Transl. Med. 8, 118 (2010).
Ma, X. et al. Identification of LIV1, a putative zinc transporter gene responsible for HDACi-induced apoptosis, using a functional gene screen approach. Mol. Cancer Ther. 8, 3108–3116 (2009).
Yamashita, S. et al. Zinc transporter LIVI controls epithelial-mesenchymal transition in zebrafish gastrula organizer. Nature 429, 298–302 (2004).
Zhao, L., Chen, W., Taylor, K.M., Cai, B. & Li, X. LIV-1 suppression inhibits HeLa cell invasion by targeting ERK1/2-Snail/Slug pathway. Biochem. Biophys. Res. Commun. 363, 82–88 (2007).
Unno, J. et al. LIV-1 enhances the aggressive phenotype through the induction of epithelial to mesenchymal transition in human pancreatic carcinoma cells. Int. J. Oncol. 35, 813–821 (2009).
Lue, H.W. et al. LIV-1 promotes prostate cancer epithelial-to-mesenchymal transition and metastasis through HB-EGF shedding and EGFR-mediated ERK signaling. PLoS ONE 6, e27720 (2011).
Lopez, V. & Kelleher, S.L. Zip6-attenuation promotes epithelial-to-mesenchymal transition in ductal breast tumor (T47D) cells. Exp. Cell Res. 316, 366–375 (2010).
Thiery, J.P. Epithelial-mesenchymal transition in tumour progression. Nat. Rev. Cancer 2, 447–454 (2002).
Jing, Y., Han, Z., Zhang, S., Liu, Y. & Wei, L. Epithelial-mesenchymal transition in tumor microenvironment. Cell Biosci. 1, 29 (2011).
Radisky, D.C. & LaBarge, M.A. Epithelial-mesenchymal transition and the stem cell phenotype. Cell Stem Cell 2, 511–512 (2008).
Taylor, K.M., Hiscox, S. & Nicholson, R.I. Zinc transporter LIV-1: a link between cellular development and cancer progression. Trends Endocrinol. Metab. 15, 461–463 (2004).
Sauna, Z.E. & Kimchi-Sarfaty, C. Understanding the contribution of synonymous mutations to human disease. Nat. Rev. Genet. 12, 683–691 (2011).
Kimchi-Sarfaty, C. et al. A “silent” polymorphism in the MDR1 gene changes substrate specificity. Science 315, 525–528 (2007).
Chen, R., Davydov, E.V., Sirota, M. & Butte, A.J. Non-synonymous and synonymous coding SNPs show similar likelihood and effect size of human disease association. PLoS ONE 5, e13574 (2010).
Wigginiton, J.E., Cutler, D.J. & Abecasis, G.R. A note on exact tests of Hardy-Weinberg equilibrium. Am. J. Hum. Genet. 76, 887–893 (2005).
Lehmann, U. & Kreipe, H. Real-time PCR analysis of DNA and RNA extracted from formalin-fixed and paraffin-embedded biopsies. Methods 25, 409–418 (2001).
Acknowledgements
We acknowledge Y. Shimada (Hyogo College of Medicine) for providing the eight ESCC cell lines. This work was funded by the National Basic Research Program of China (2013CB910303 and 2011CB504303) and the National Natural Science Foundation of China (91229126).
Author information
Authors and Affiliations
Contributions
Dongxin Lin was the overall principal investigator of the study who conceived the study, obtained financial support, was responsible for study design, oversaw the entire study, interpreted the results, and wrote parts of and synthesized the manuscript. C.W. performed overall project management, oversaw laboratory analyses, performed statistical analyses and drafted the initial version of the manuscript. D. Li, Y.Q., Yuling Zhou, J.C., K.Z. and Menghan Wang performed functional analyses. W.J. and Y. Zeng were responsible for subject recruitment and sample preparation for Guangzhou samples. Z.H., Yifeng Zhou and H.S. were responsible for subject recruitment and sample preparation for Jiangsu samples. D.Y. and W.T. performed subject recruitment and sample preparation for Beijing samples. T.T. and Mingrong Wang were responsible for subject recruitment and preparation of ESCC tissue microarrays. Dongmei Lin and L.W. were responsible for the IHC analysis of tissue microarrays.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Text and Figures
Supplementary Tables 1 and 3, Supplementary Figures 1–6 (PDF 1212 kb)
Supplementary Table 2
Associations of SNPs in SLC39A6 region with hazard risk of death of ESCC patients in the discovery sample (XLSX 13 kb)
Rights and permissions
About this article
Cite this article
Wu, C., Li, D., Jia, W. et al. Genome-wide association study identifies common variants in SLC39A6 associated with length of survival in esophageal squamous-cell carcinoma. Nat Genet 45, 632–638 (2013). https://doi.org/10.1038/ng.2638
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ng.2638
This article is cited by
-
The association between depression and esophageal cancer in China: a multicentre population-based study
BMC Psychiatry (2021)
-
Intronic polymorphisms in genes LRFN2 (rs2494938) and DNAH11 (rs2285947) are prognostic indicators of esophageal squamous cell carcinoma
BMC Medical Genetics (2019)
-
Involvement of SLC39A6 in gastric adenocarcinoma and correlation of the SLC39A6 polymorphism rs1050631 with clinical outcomes after resection
BMC Cancer (2019)
-
Maintenance of Intestinal Epithelial Homeostasis by Zinc Transporters
Digestive Diseases and Sciences (2019)
-
Epigenetic silencing of ZNF132 mediated by methylation-sensitive Sp1 binding promotes cancer progression in esophageal squamous cell carcinoma
Cell Death & Disease (2018)