Abstract
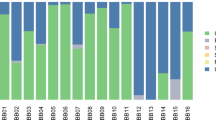
The importance of commensal microbes for human health is increasingly recognized1,2,3,4,5, yet the impacts of evolutionary changes in human diet and culture on commensal microbiota remain almost unknown. Two of the greatest dietary shifts in human evolution involved the adoption of carbohydrate-rich Neolithic (farming) diets6,7 (beginning ∼10,000 years before the present6,8) and the more recent advent of industrially processed flour and sugar (in ∼1850)9. Here, we show that calcified dental plaque (dental calculus) on ancient teeth preserves a detailed genetic record throughout this period. Data from 34 early European skeletons indicate that the transition from hunter-gatherer to farming shifted the oral microbial community to a disease-associated configuration. The composition of oral microbiota remained unexpectedly constant between Neolithic and medieval times, after which (the now ubiquitous) cariogenic bacteria became dominant, apparently during the Industrial Revolution. Modern oral microbiotic ecosystems are markedly less diverse than historic populations, which might be contributing to chronic oral (and other) disease in postindustrial lifestyles.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
Accession codes
References
Aas, J.A. et al. Bacteria of dental caries in primary and permanent teeth in children and young adults. J. Clin. Microbiol. 46, 1407–1417 (2008).
Aas, J.A., Paster, B.J., Stokes, L.N., Olsen, I. & Dewhirst, F.E. Defining the normal bacterial flora of the oral cavity. J. Clin. Microbiol. 43, 5721–5732 (2005).
Davé, S. & Van Dyke, T. The link between periodontal disease and cardiovascular disease is probably inflammation. Oral Dis. 14, 95–101 (2008).
Grossi, S.G. & Genco, R.J. Periodontal disease and diabetes mellitus: a two-way relationship. Ann. Periodontol. 3, 51–61 (1998).
Human Microbiome Project Consortium. Structure, function and diversity of the healthy human microbiome. Nature 486, 207–214 (2012).
Braidwood, R.J., Howe, B. & Reed, C.A. The Iranian Prehistoric Project: new problems arise as more is learned of the first attempts at food production and settled village life. Science 133, 2008–2010 (1961).
Oelzea, V.M. et al. Early Neolithic diet and animal husbandry: stable isotope evidence from three Linearbandkeramik (LBK) sites in Central Germany. J. Archaeol. Sci. 38, 270–279 (2011).
Childe, V.G. The Dawn of European Civilisation (Kegan Paul, London, 1925).
Cordain, L. et al. Origins and evolution of the Western diet: health implications for the 21st century. Am. J. Clin. Nutr. 81, 341–354 (2005).
Savage, D.C. Microbial ecology of the gastrointestinal tract. Annu. Rev. Microbiol. 31, 107–133 (1977).
Scott, G.R. & Poulson, S.R. Stable carbon and nitrogen isotopes of human dental calculus: a potentially new non-destructive proxy for paleodietary analysis. J. Archaeol. Sci. 39, 1388–1393 (2012).
Lilley, J., Stroud, G. & Brothwell, D. The Jewish burial ground at Jewbury. in The Archaeology of York Vol. 12 (eds. Addyman, P.V. & Kinsler, V.A.) 291–578 (Council for British Archaeology, York, UK, 1994).
Preus, H.R., Marvik, O.J., Selvig, K.A. & Bennike, P. Ancient bacterial DNA (aDNA) in dental calculus from archaeological human remains. J. Archaeol. Sci. 38, 1827–1831 (2011).
Vandermeersch, B. et al. Middle Palaeolithic dental bacteria from Kebara, Israel. C.R. Acad. Sci. Paris 319, 727–731 (1994).
Socransky, S.S. & Haffajee, A.D. Dental biofilms: difficult therapeutic targets. Periodontol. 2000 28, 12–55 (2002).
Jin, Y. & Yip, H.K. Supragingival calculus: formation and control. Crit. Rev. Oral Biol. Med. 13, 426–441 (2002).
Lieverse, A.R. Diet and the aetiology of dental calculus. Int. J. Osteoarchaeol. 9, 219–232 (1999).
Asikainen, S., Chen, C. & Slots, J. Likelihood of transmitting Actinobacillus actinomycetemcomitans and Porphyromonas gingivalis in families with periodontitis. Oral Microbiol. Immunol. 11, 387–394 (1996).
Van Steenbergen, T.J., Menard, C., Tijhof, C.J., Mouton, C. & De Graaff, J. Comparison of three molecular typing methods in studies of transmission of Porphyromonas gingivalis. J. Med. Microbiol. 39, 416–421 (1993).
Aufderheide, A.C., Rodriguez-Martin, C. & Langsjoen, O. The Cambridge Encyclopedia of Human Paleopathology (Cambridge University Press, Cambridge, 1998).
Faveri, M. et al. Microbiological diversity of generalized aggressive periodontitis by 16S rRNA clonal analysis. Oral Microbiol. Immunol. 23, 112–118 (2008).
Grine, F.E., Gwinnett, A.J. & Oaks, J.H. Early hominid dental pathology: interproximal caries in 1.5 million-year-old Paranthropus robustus from Swartkrans. Arch. Oral Biol. 35, 381–386 (1990).
Marsh, P.D. Sugar, fluoride, pH and microbial homeostasis in dental plaque. Proc. Finn. Dent. Soc. 87, 515–525 (1991).
Marsh, P.D. Are dental diseases examples of ecological catastrophes? Microbiology 149, 279–294 (2003).
Hujoel, P. Dietary carbohydrates and dental-systemic diseases. J. Dent. Res. 88, 490–502 (2009).
Marsh, P.D. Microbiology of dental plaque biofilms and their role in oral health and caries. Dent. Clin. North Am. 54, 441–454 (2010).
Petersen, P.E., Bourgeois, D., Ogawa, H., Estupinan-Day, S. & Ndiaye, C. The global burden of oral diseases and risks to oral health. Bull. World Health Organ. 83, 661–669 (2005).
Mercado, F.B., Marshall, R.I., Klestov, A.C. & Bartold, P.M. Relationship between rheumatoid arthritis and periodontitis. J. Periodontol. 72, 779–787 (2001).
Quince, C. et al. Accurate determination of microbial diversity from 454 pyrosequencing data. Nat. Methods 6, 639–641 (2009).
Dewhirst, F.E. et al. The human oral microbiome. J. Bacteriol. 192, 5002–5017 (2010).
Lazarevic, V., Whiteson, K., Hernandez, D., Francois, P. & Schrenzel, J. Study of inter- and intra-individual variations in the salivary microbiota. BMC Genomics 11, 523 (2010).
La Duc, M.T., Kern, R. & Venkateswaran, K. Microbial monitoring of spacecraft and associated environments. Microb. Ecol. 47, 150–158 (2004).
D'Costa, V.M. et al. Antibiotic resistance is ancient. Nature 477, 457–461 (2011).
Beier, S., Witzel, K.P. & Marxsen, J. Bacterial community composition in Central European running waters examined by temperature gradient gel electrophoresis and sequence analysis of 16S rRNA genes. Appl. Environ. Microbiol. 74, 188–199 (2008).
Schloss, P.D. & Handelsman, J. Toward a census of bacteria in soil. PLoS Comput. Biol. 2, e92 (2006).
Ellis, R.J., Morgan, P., Weightman, A.J. & Fry, J.C. Cultivation-dependent and -independent approaches for determining bacterial diversity in heavy-metal-contaminated soil. Appl. Environ. Microbiol. 69, 3223–3230 (2003).
Nogales, B. et al. Combined use of 16S ribosomal DNA and 16S rRNA to study the bacterial community of polychlorinated biphenyl–polluted soil. Appl. Environ. Microbiol. 67, 1874–1884 (2001).
Elshahed, M.S. et al. Novelty and uniqueness patterns of rare members of the soil biosphere. Appl. Environ. Microbiol. 74, 5422–5428 (2008).
Tringe, S.G. et al. Comparative metagenomics of microbial communities. Science 308, 554–557 (2005).
Will, C. et al. Horizon-specific bacterial community composition of German grassland soils, as revealed by pyrosequencing-based analysis of 16S rRNA genes. Appl. Environ. Microbiol. 76, 6751–6759 (2010).
Kerr, N.W. Prevalence and natural history of periodontal disease in prehistoric Scots (pre-900 AD). J. Periodontal Res. 33, 131–137 (1998).
Albandar, J.M., Brunelle, J.A. & Kingman, A. Destructive periodontal disease in adults 30 years of age and older in the United States, 1988–1994. J. Periodontol. 70, 13–29 (1999).
Bailey, M.T. et al. Stressor exposure disrupts commensal microbial populations in the intestines and leads to increased colonization by Citrobacter rodentium. Infect. Immun. 78, 1509–1519 (2010).
Lawley, T.D. et al. Antibiotic treatment of Clostridium difficile carrier mice triggers a supershedder state, spore-mediated transmission, and severe disease in immunocompromised hosts. Infect. Immun. 77, 3661–3669 (2009).
Lozupone, C.A., Stombaugh, J.I., Gordon, J.I., Jansson, J.K. & Knight, R. Diversity, stability and resilience of the human gut microbiota. Nature 489, 220–230 (2012).
Cadotte, M., Dinnage, R. & Tilman, G.D. Phylogenetic diversity promotes ecosystem stability. Ecology 93, S223–S233 (2012).
Zhang, Y., Chen, H.Y.H. & Reich, P.B. Forest productivity increases with evenness, species richness and trait variation: a global meta-analysis. J. Ecol. 100, 742–749 (2012).
Petchey, O. & Gaston, K. Effects on ecosystem resilience of biodiversity, extinctions, and the structure of regional species pools. Theor. Ecol. 2, 177–187 (2009).
Loreau, M. et al. A new look at the relationship between diversity and stability. in Biodiversity and Ecosystem Functioning. Synthesis and Perspectives (eds. Loreau, M., Naeem, S. & Inchausti, P.) 79–91 (Oxford University Press, Oxford, 2002).
Faith, D.P. Conservation evaluation and phylogenetic diversity. Biol. Conserv. 61, 1–10 (1992).
Haak, W. et al. Ancient DNA from European early neolithic farmers reveals their near eastern affinities. PLoS Biol. 8, e1000536 (2010).
Caporaso, J.G. et al. QIIME allows analysis of high-throughput community sequencing data. Nat. Methods 7, 335–336 (2010).
Reeder, J. & Knight, R. Rapidly denoising pyrosequencing amplicon reads by exploiting rank-abundance distributions. Nat. Methods 7, 668–669 (2010).
Edgar, R.C. Search and clustering orders of magnitude faster than BLAST. Bioinformatics 26, 2460–2461 (2010).
Schloss, P.D. The effects of alignment quality, distance calculation method, sequence filtering, and region on the analysis of 16S rRNA gene-based studies. PLoS Comput. Biol. 6, e1000844 (2010).
Weisburg, W.G., Barns, S.M., Pelletier, D.A. & Lane, D.J. 16S ribosomal DNA amplification for phylogenetic study. J. Bacteriol. 173, 697–703 (1991).
Wang, Q., Garrity, G.M., Tiedje, J.M. & Cole, J.R. Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl. Environ. Microbiol. 73, 5261–5267 (2007).
Acknowledgements
We thank D. Brothwell for original inspiration, N. Gully and S. Bent for critical discussions and J. Soubrier for bioinformatics assistance. We thank H. Meller from the State Heritage Museum of Saxony-Anhalt, Germany, and W. Gumiński from the Institute of Archaeology, University of Warsaw, Poland, for prehistoric samples and members of the Australian Centre for Ancient DNA for practical help and providing samples of plaque and calculus. We thank several anonymous reviewers whose comments have considerably improved the manuscript. We thank the Australian Research Council, the Wellcome Trust (WT092799/Z/10/Z and WT098051) and the Sir Mark Mitchell Foundation for funding support.
Author information
Authors and Affiliations
Contributions
C.J.A., A.C., K.D., A.W.W., J.P., K.W.A., G.T., J.K. and W.H. designed the study. C.J.A., K.D., K.W.A., A.S., W.H., A.C. and J.K. collected samples. C.J.A. and L.S.W. extracted and amplified DNA from dental calculus. C.J.A. and L.S.W. analyzed sequence data. A.W.W. performed 454 sequencing. C.J.A.B. performed α diversity bootstrapping analyses. C.J.A., A.C. and K.D. wrote the manuscript. All authors discussed the results and contributed to writing the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–11, Supplementary Tables 1–9 and Supplementary Note (PDF 1179 kb)
Rights and permissions
About this article
Cite this article
Adler, C., Dobney, K., Weyrich, L. et al. Sequencing ancient calcified dental plaque shows changes in oral microbiota with dietary shifts of the Neolithic and Industrial revolutions. Nat Genet 45, 450–455 (2013). https://doi.org/10.1038/ng.2536
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ng.2536
This article is cited by
-
Identification of microbial pathogens in Neolithic Scandinavian humans
Scientific Reports (2024)
-
Evolutionary History of Periodontitis and the Oral Microbiota—Lessons for the Future
Current Oral Health Reports (2024)
-
Ancient dental calculus reveals oral microbiome shifts associated with lifestyle and disease in Great Britain
Nature Microbiology (2023)
-
Salivary bacterial signatures in depression-obesity comorbidity are associated with neurotransmitters and neuroactive dipeptides
BMC Microbiology (2022)
-
Dental calculus - oral health, forensic studies and archaeology: a review
British Dental Journal (2022)