Abstract
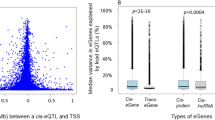
Identifying the downstream effects of disease-associated SNPs is challenging. To help overcome this problem, we performed expression quantitative trait locus (eQTL) meta-analysis in non-transformed peripheral blood samples from 5,311 individuals with replication in 2,775 individuals. We identified and replicated trans eQTLs for 233 SNPs (reflecting 103 independent loci) that were previously associated with complex traits at genome-wide significance. Some of these SNPs affect multiple genes in trans that are known to be altered in individuals with disease: rs4917014, previously associated with systemic lupus erythematosus (SLE)1, altered gene expression of C1QB and five type I interferon response genes, both hallmarks of SLE2,3,4. DeepSAGE RNA sequencing showed that rs4917014 strongly alters the 3′ UTR levels of IKZF1 in cis, and chromatin immunoprecipitation and sequencing analysis of the trans-regulated genes implicated IKZF1 as the causal gene. Variants associated with cholesterol metabolism and type 1 diabetes showed similar phenomena, indicating that large-scale eQTL mapping provides insight into the downstream effects of many trait-associated variants.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
Accession codes
Primary accessions
ArrayExpress
Gene Expression Omnibus
References
Han, J.W. et al. Genome-wide association study in a Chinese Han population identifies nine new susceptibility loci for systemic lupus erythematosus. Nat. Genet. 41, 1234–1237 (2009).
Bengtsson, A.A. et al. Activation of type I interferon system in systemic lupus erythematosus correlates with disease activity but not with antiretroviral antibodies. Lupus 9, 664–671 (2000).
Bohlson, S.S., Fraser, D.A. & Tenner, A.J. Complement proteins C1q and MBL are pattern recognition molecules that signal immediate and long-term protective immune functions. Mol. Immunol. 44, 33–43 (2007).
Ytterberg, S.R. & Schnitzer, T.J. Serum interferon levels in patients with systemic lupus erythematosus. Arthritis Rheum. 25, 401–406 (1982).
Fehrmann, R.S. et al. Trans-eQTLs reveal that independent genetic variants associated with a complex phenotype converge on intermediate genes, with a major role for the HLA. PLoS Genet. 7, e1002197 (2011).
Nicolae, D.L. et al. Trait-associated SNPs are more likely to be eQTLs: annotation to enhance discovery from GWAS. PLoS Genet. 6, e1000888 (2010).
Pickrell, J.K. et al. Understanding mechanisms underlying human gene expression variation with RNA sequencing. Nature 464, 768–772 (2010).
Dubois, P.C. et al. Multiple common variants for celiac disease influencing immune gene expression. Nat. Genet. 42, 295–302 (2010).
Fairfax, B.P. et al. Genetics of gene expression in primary immune cells identifies cell type–specific master regulators and roles of HLA alleles. Nat. Genet. 44, 502–510 (2012).
Innocenti, F. et al. Identification, replication, and functional fine-mapping of expression quantitative trait loci in primary human liver tissue. PLoS Genet. 7, e1002078 (2011).
Grundberg, E. et al. Mapping cis- and trans-regulatory effects across multiple tissues in twins. Nat. Genet. 44, 1084–1089 (2012).
Heinig, M. et al. A trans-acting locus regulates an anti-viral expression network and type 1 diabetes risk. Nature 467, 460–464 (2010).
Small, K.S. et al. Identification of an imprinted master trans regulator at the KLF14 locus related to multiple metabolic phenotypes. Nat. Genet. 43, 561–564 (2011).
Metspalu, A. The Estonian Genome Project. Drug Dev. Res. 62, 97–101 (2004).
Tanaka, T. et al. Genome-wide association study of plasma polyunsaturated fatty acids in the InCHIANTI Study. PLoS Genet. 5, e1000338 (2009).
Hofman, A. et al. The Rotterdam Study: 2012 objectives and design update. Eur. J. Epidemiol. 26, 657–686 (2011).
Heckbert, S.R. et al. Antihypertensive treatment with ACE inhibitors or β-blockers and risk of incident atrial fibrillation in a general hypertensive population. Am. J. Hypertens. 22, 538–544 (2009).
Psaty, B.M. et al. The risk of myocardial infarction associated with antihypertensive drug therapies. J. Am. Med. Assoc. 274, 620–625 (1995).
Smith, N.L. et al. Esterified estrogens and conjugated equine estrogens and the risk of venous thrombosis. J. Am. Med. Assoc. 292, 1581–1587 (2004).
Teumer, A. et al. Genome-wide association study identifies four genetic loci associated with thyroid volume and goiter risk. Am. J. Hum. Genet. 88, 664–673 (2011).
Inouye, M. et al. An immune response network associated with blood lipid levels. PLoS Genet. 6, e1001113 (2010).
Mehta, D. et al. Impact of common regulatory single-nucleotide variants on gene expression profiles in whole blood. Eur. J. Hum. Genet. 21, 48–54 (2013).
Powell, J.E. et al. The Brisbane Systems Genetics Study: genetical genomics meets complex trait genetics. PLoS ONE 7, e35430 (2012).
Wang, C. et al. Genes identified in Asian SLE GWASs are also associated with SLE in Caucasian populations. Eur. J. Hum. Genet. 10.1038/ejhg.2012.277 (19 December 2012).
1000 Genomes Project Consortium. A map of human genome variation from population-scale sequencing. Nature 467, 1061–1073 (2010).
Patterson, K. 1000 genomes: a world of variation. Circ. Res. 108, 534–536 (2011).
Baechler, E.C. et al. Interferon-inducible gene expression signature in peripheral blood cells of patients with severe lupus. Proc. Natl. Acad. Sci. USA 100, 2610–2615 (2003).
Bennett, L. et al. Interferon and granulopoiesis signatures in systemic lupus erythematosus blood. J. Exp. Med. 197, 711–723 (2003).
McAdam, R.A., Goundis, D. & Reid, K.B. A homozygous point mutation results in a stop codon in the C1q B-chain of a C1q-deficient individual. Immunogenetics 27, 259–264 (1988).
Botto, M. et al. Homozygous C1q deficiency causes glomerulonephritis associated with multiple apoptotic bodies. Nat. Genet. 19, 56–59 (1998).
Zhernakova, D.V. et al. DeepSAGE reveals genetic variants associated with alternative polyadenylation and expression of coding and non-coding transcripts. PLoS Genet. 9, e1003594 (2013).
ENCODE Project Consortium. An integrated encyclopedia of DNA elements in the human genome. Nature 489, 57–74 (2012).
Ganesh, S.K. et al. Multiple loci influence erythrocyte phenotypes in the CHARGE Consortium. Nat. Genet. 41, 1191–1198 (2009).
Wang, J.H. et al. Selective defects in the development of the fetal and adult lymphoid system in mice with an Ikaros null mutation. Immunity 5, 537–549 (1996).
Zabaneh, D. & Balding, D.J. A genome-wide association study of the metabolic syndrome in Indian Asian men. PLoS ONE 5, e11961 (2010).
Sabatti, C. et al. Genome-wide association analysis of metabolic traits in a birth cohort from a founder population. Nat. Genet. 41, 35–46 (2009).
Teslovich, T.M. et al. Biological, clinical and population relevance of 95 loci for blood lipids. Nature 466, 707–713 (2010).
Barrett, J.C. et al. Genome-wide association study and meta-analysis find that over 40 loci affect risk of type 1 diabetes. Nat. Genet. 41, 703–707 (2009).
Plagnol, V. et al. Genome-wide association analysis of autoantibody positivity in type 1 diabetes cases. PLoS Genet. 7, e1002216 (2011).
Zhernakova, A. et al. Meta-analysis of genome-wide association studies in celiac disease and rheumatoid arthritis identifies fourteen non-HLA shared loci. PLoS Genet. 7, e1002004 (2011).
Eriksson, N. et al. Novel associations for hypothyroidism include known autoimmune risk loci. PLoS ONE 7, e34442 (2012).
Jin, Y. et al. Genome-wide association analyses identify 13 new susceptibility loci for generalized vitiligo. Nat. Genet. 44, 676–680 (2012).
Newton-Cheh, C. et al. Genome-wide association study identifies eight loci associated with blood pressure. Nat. Genet. 41, 666–676 (2009).
Wain, L.V. et al. Genome-wide association study identifies six new loci influencing pulse pressure and mean arterial pressure. Nat. Genet. 43, 1005–1011 (2011).
Köttgen, A. et al. New loci associated with kidney function and chronic kidney disease. Nat. Genet. 42, 376–384 (2010).
Gudbjartsson, D.F. et al. Sequence variants affecting eosinophil numbers associate with asthma and myocardial infarction. Nat. Genet. 41, 342–347 (2009).
Rotival, M. et al. Integrating genome-wide genetic variations and monocyte expression data reveals trans-regulated gene modules in humans. PLoS Genet. 7, e1002367 (2011).
International HapMap Consortium. The International HapMap Project. Nature 426, 789–796 (2003).
Westra, H.J. et al. MixupMapper: correcting sample mix-ups in genome-wide datasets increases power to detect small genetic effects. Bioinformatics 27, 2104–2111 (2011).
Breitling, R. et al. Genetical genomics: spotlight on QTL hotspots. PLoS Genet. 4, e1000232 (2008).
Whitlock, M.C. Combining probability from independent tests: the weighted Z-method is superior to Fisher's approach. J. Evol. Biol. 18, 1368–1373 (2005).
Alberts, R. et al. Sequence polymorphisms cause many false cis eQTLs. PLoS ONE 2, e622 (2007).
Benovoy, D., Kwan, T. & Majewski, J. Effect of polymorphisms within probe-target sequences on olignonucleotide microarray experiments. Nucleic Acids Res. 36, 4417–4423 (2008).
Xu, Z. & Taylor, J.A. SNPinfo: integrating GWAS and candidate gene information into functional SNP selection for genetic association studies. Nucleic Acids Res. 37, W600–W605 (2009).
Dayem Ullah, A.Z., Lemoine, N.R. & Chelala, C. SNPnexus: a web server for functional annotation of novel and publicly known genetic variants (2012 update). Nucleic Acids Res. 40, W65–W70 (2012).
Chelala, C., Khan, A. & Lemoine, N.R. SNPnexus: a web database for functional annotation of newly discovered and public domain single nucleotide polymorphisms. Bioinformatics 25, 655–661 (2009).
Ward, L.D. & Kellis, M. HaploReg: a resource for exploring chromatin states, conservation, and regulatory motif alterations within sets of genetically linked variants. Nucleic Acids Res. 40, D930–D934 (2012).
Flicek, P. et al. Ensembl 2012. Nucleic Acids Res. 40, D84–D90 (2012).
Hindorff, L.A. et al. Potential etiologic and functional implications of genome-wide association loci for human diseases and traits. Proc. Natl. Acad. Sci. USA 106, 9362–9367 (2009).
Acknowledgements
Acknowledgments for each participating cohort can be found in the Supplementary Note.
Author information
Authors and Affiliations
Contributions
Experiment design and method development: H.-J.W., M.J.P., T.E., H.Y., C.S., J. Kettunen, M.W.C., B.P.F., K. Schramm, J. Karjalainen, T.L., Y.L., R.C.J., B.M.P., S.R., A.T., T.M.F., A.M., J.B.J.M. and L. Franke. Reviewing and editing of the manuscript: H.-J.W., M.J.P., T.E., H.Y., C.S., J. Kettunen, M.W.C., B.P.F., A.Z., A.G.U., A.H., F.R., V.S., J.B., T.L., Y.L., R.C.J., P.M.V., J.C.K., B.M.P., S.R., A.T., T.M.F., A.M., J.B.J.M. and L. Franke. Data collection: D.V.Z., J.H.V., J. Karjalainen, S.W., F.R., P.A.C.t.H., E.R., K.F., M. Nelis, L.M., D.M., L. Ferrucci, A.B.S., D.G.H., M.A.N., G.H., M. Nauck, D.R., U.V., M.P., A.S.-D., S.A.G., D.A.E., G.W.M., S.M., H.P., C.H., M.R., H.G., T.M., K. Strauch and L.H.V.d.B. Replication of trans-eQTL results: B.P.F., K. Schramm, J.E.P., P.M.V. and J.C.K.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–18, Supplementary Tables 1–10 and Supplementary Note (PDF 48998 kb)
Source data
Rights and permissions
About this article
Cite this article
Westra, HJ., Peters, M., Esko, T. et al. Systematic identification of trans eQTLs as putative drivers of known disease associations. Nat Genet 45, 1238–1243 (2013). https://doi.org/10.1038/ng.2756
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ng.2756
This article is cited by
-
Genome-wide identification of RNA modification-related single nucleotide polymorphisms associated with rheumatoid arthritis
BMC Genomics (2023)
-
Exploring the novel SNPs in neuroticism and birth weight based on GWAS datasets
BMC Medical Genomics (2023)
-
Evaluating 17 methods incorporating biological function with GWAS summary statistics to accelerate discovery demonstrates a tradeoff between high sensitivity and high positive predictive value
Communications Biology (2023)
-
Genome-wide association study of a lipedema phenotype among women in the UK Biobank identifies multiple genetic risk factors
European Journal of Human Genetics (2023)
-
Molecular pathways identified from single nucleotide polymorphisms demonstrate mechanistic differences in systemic lupus erythematosus patients of Asian and European ancestry
Scientific Reports (2023)