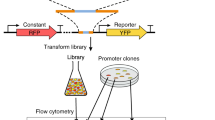
Abstract
The precise tuning of gene expression levels is essential for the optimal performance of transcriptional regulatory networks. We created 209 variants of the Saccharomyces cerevisiae PHO5 promoter to quantify how different binding sites for the transcription factor Pho4 affect its output. We found that transcription-factor binding affinities determined in vitro could quantitatively predict the output of a complex yeast promoter. Promoter output was precisely tunable by subtle changes in binding-site affinity of less than 3 kcal mol−1, which are accessible by modifying 1–2 bases. Our results provide insights into how transcription-factor binding sites regulate gene expression, their possible evolution and how they can be used to precisely tune gene expression. More generally, we show that in vitro binding-energy landscapes of transcription factors can precisely predict the output of a native yeast promoter, indicating that quantitative models of transcriptional regulatory networks are feasible.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout








Similar content being viewed by others
References
Segal, E. & Widom, J. From DNA sequence to transcriptional behaviour: a quantitative approach. Nat. Rev. Genet. 10, 443–456 (2009).
Geertz, M. & Maerkl, S.J. Experimental strategies for studying transcription factor–DNA binding specificities. Brief Funct. Genomics 9, 362–373 (2010).
Gertz, J., Siggia, E.D. & Cohen, B.A. Analysis of combinatorial cis-regulation in synthetic and genomic promoters. Nature 457, 215–218 (2009).
Patwardhan, R.P. et al. High-resolution analysis of DNA regulatory elements by synthetic saturation mutagenesis. Nat. Biotechnol. 27, 1173–1175 (2009).
Kwasnieski, J.C., Mogno, I., Myers, C.A., Corbo, J.C. & Cohen, B.A. Complex effects of nucleotide variants in a mammalian cis-regulatory element. Proc. Natl. Acad. Sci. USA 109, 19498–19503 (2012).
Zeevi, D. et al. Compensation for differences in gene copy number among yeast ribosomal proteins is encoded within their promoters. Genome Res. 21, 2114–2128 (2011).
Kinney, J.B., Murugan, A., Callan, C.G. Jr. & Cox, E.C. Using deep sequencing to characterize the biophysical mechanism of a transcriptional regulatory sequence. Proc. Natl. Acad. Sci. USA 107, 9158–9163 (2010).
Cox, R.S. III, Surette, M.G. & Elowitz, M.B. Programming gene expression with combinatorial promoters. Mol. Syst. Biol. 3, 145 (2007).
Gertz, J. & Cohen, B.A. Environment-specific combinatorial cis-regulation in synthetic promoters. Mol. Syst. Biol. 5, 9 (2009).
Patwardhan, R.P. et al. Massively parallel functional dissection of mammalian enhancers in vivo. Nat. Biotechnol. 30, 265–270 (2012).
Melnikov, A. et al. Systematic dissection and optimization of inducible enhancers in human cells using a massively parallel reporter assay. Nat. Biotechnol. 30, 271–277 (2012).
Lam, F.H., Steger, D.J. & O'Shea, E.K. Chromatin decouples promoter threshold from dynamic range. Nature 458, 6 (2008).
Aow, J.S. et al. Differential binding of the related transcription factors Pho4 and Cbf1 can tune the sensitivity of promoters to different levels of an induction signal. Nucleic Acids Res. 41, 4877–4887 (2013).
Sharon, E. et al. Inferring gene regulatory logic from high-throughput measurements of thousands of systematically designed promoters. Nat. Biotechnol. 30, 521–530 (2012).
Tanay, A. Extensive low-affinity transcriptional interactions in the yeast genome. Genome Res. 16, 962–972 (2006).
Kahana, S. et al. Functional dissection of IME1 transcription using quantitative promoter-reporter screening. Genetics 186, 829–841 (2010).
Su, T.C., Tamarkina, E. & Sadowski, I. Organizational constraints on Ste12 cis-elements for a pheromone response in Saccharomyces cerevisiae. FEBS J. 277, 3235–3248 (2010).
Khalil, A.S. et al. A synthetic biology framework for programming eukaryotic transcription functions. Cell 150, 647–658 (2012).
Raveh-Sadka, T. et al. Manipulating nucleosome disfavoring sequences allows fine-tune regulation of gene expression in yeast. Nat. Genet. 44, 743–750 (2012).
Svaren, J. & Hörz, W. Transcription factors vs nucleosomes: regulation of the PHO5 promoter in yeast. Trends Biochem. Sci. 22, 5 (1997).
Maerkl, S.J. & Quake, S.R. A systems approach to measuring the binding energy landscapes of transcription factors. Science 315, 233–237 (2007).
Oshima, Y. The phosphatase system in Saccharomyces cerevisae. Genes Genet. Syst. 72, 323–334 (1997).
Ogawa, N., DeRisi, J.L. & Brown, P.O. New components of a system for phosphate accumulation and polyphosphate metabolism in Saccharomyces cerevisiae revealed by genomic expression analysis. Mol. Biol. Cell 11, 13–22 (2000).
Venter, U., Svaren, J., Schmitz, J., Schmid, A. & Hörz, W. A nucleosome precludes binding of the transcription factor Pho4 in vivo to a critical target site in the PHO5 promoter. EMBO J. 13, 8 (1994).
Barbarić, S., Münsterkötter, M., Goding, C.R. & Hörz, W. Cooperative Pho2-Pho4 interactions at the PHO5 promoter are critical for binding of Pho4 to UASp1 and for efficient transactivation by Pho4 at UASp2. Mol. Cell Biol. 18, 11 (1998).
Barbarić, S., Münsterkötter, M., Svaren, J. & Hörz, W. The homeodomain protein Pho2 and the basic-helix-loop-helix protein Pho4 bind DNA cooperatively at the yeast PHO5 promoter. Nucleic Acids Res. 24, 4479–4486 (1996).
Rajkumar, A.S. & Maerkl, S.J. Rapid synthesis of defined eukaryotic promoter libraries. ACS Synth. Biol. 1, 483–490 (2012).
Lee, W. et al. A high-resolution atlas of nucleosome occupancy in yeast. Nat. Genet. 39, 1235–1244 (2007).
Shimizu, T. et al. Crystal structure of PHO4 bHLH domain–DNA complex: flanking base recognition. EMBO J. 16, 4689–4697 (1997).
Fisher, F. & Goding, C.R. Single amino acid substitutions alter helix-loop-helix protein specificity for bases flanking the core CANNTG motif. EMBO J. 11, 7 (1992).
Maerkl, S.J. & Quake, S.R. Experimental determination of the evolvability of a transcription factor. Proc. Natl. Acad. Sci. USA 106, 18650–18655 (2009).
Mogno, I., Vallania, F., Mitra, R.D. & Cohen, B.A. TATA is a modular component of synthetic promoters. Genome Res. 20, 1391–1397 (2010).
Granek, J.A. & Clarke, N.D. Explicit equilibrium modeling of transcription-factor binding and gene regulation. Genome Biol. 6, R87 (2005).
Bintu, L. et al. Transcriptional regulation by the numbers: models. Curr. Opin. Genet. Dev. 15, 116–124 (2005).
Zhou, X. & O'Shea, E.K. Integrated approaches reveal determinants of genome-wide binding and function of the transcription factor Pho4. Mol. Cell 42, 826–836 (2011).
Mahony, S. & Benos, P.V. STAMP: a web tool for exploring DNA-binding motif similarities. Nucleic Acids Res. 35, W253–W258 (2007).
Spivak, A.T. & Stormo, G.D. ScerTF: a comprehensive database of benchmarked position weight matrices for Saccharomyces species. Nucleic Acids Res. 40, D162–D168 (2012).
Wykoff, D.D. & O'Shea, E.K. Phosphate transport and sensing in Saccharomyces cerevisiae. Genetics 159, 10 (2001).
Springer, M., Wykoff, D.D., Miller, N. & O'Shea, E.K. Partially phosphorylated Pho4 activates transcription of a subset of phosphate-responsive genes. PLoS Biol. 1, E28 (2003).
Raveh-Sadka, T., Levo, M. & Segal, E. Incorporating nucleosomes into thermodynamic models of transcription regulation. Genome Res. 19, 1480–1496 (2009).
Chiang, D.Y., Nix, D.A., Shultzaberger, R.K., Gasch, A.P. & Eisen, M.B. Flexible promoter architecture requirements for coactivator recruitment. BMC Mol. Biol. 7, 16 (2006).
Janssens, H. et al. Quantitative and predictive model of transcriptional control of the Drosophila melanogaster even skipped gene. Nat. Genet. 38, 1159–1165 (2006).
Kim, H.D. & O'Shea, E.K. A quantitative model of transcription factor-activated gene expression. Nat. Struct. Mol. Biol. 15, 1192–1198 (2008).
Mao, C. et al. Quantitative analysis of the transcription control mechanism. Mol. Syst. Biol. 6, 431 (2010).
Gietz, R.D. & Schiestl, R.H. High-efficiency yeast transformation using the LiAc/SS carrier DNA/PEG method. Nat. Protoc. 2, 31–34 (2007).
MacIsaac, K.D. et al. An improved map of conserved regulatory sites for Saccharomyces cerevisiae. BMC Bioinformatics 7, 113 (2006).
Zhu, C. et al. High-resolution DNA-binding specificity analysis of yeast transcription factors. Genome Res. 19, 556–566 (2009).
Sheff, M.A. & Thorn, K.S. Optimized cassettes for fluorescent protein tagging in Saccharomyces cerevisiae. Yeast 21, 661–670 (2004).
Schneider, T.D. Information content of individual genetic sequences. J. Theor. Biol. 189, 427–441 (1997).
Schneider, T.D. & Stephens, R.M. Sequence logos:a new way to display consensus sequences. Nucleic Acids Res. 18, 6097–6100 (1990).
Wang, Y. et al. Precision and functional specificity in mRNA decay. Proc. Natl. Acad. Sci. USA 99, 5860–5865 (2002).
Miller, C. et al. Dynamic transcriptome analysis measures rates of mRNA synthesis and decay in yeast. Mol. Syst. Biol. 7, 458 (2011).
Belle, A., Tanay, A., Bitincka, L., Shamir, R. & O'Shea, E.K. Quantification of protein half-lives in the budding yeast proteome. Proc. Natl. Acad. Sci. USA 103, 13004–13009 (2006).
Karpinets, T.V., Greenwood, D.J., Sams, C.E. & Ammons, J.T. RNA:protein ratio of the unicellular organism as a characteristic of phosphorous and nitrogen stoichiometry and of the cellular requirement of ribosomes for protein synthesis. BMC Biol. 4, 30 (2006).
Arava, Y. et al. Genome-wide analysis of mRNA translation profiles in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 100, 3889–3894 (2003).
Shaner, N.C. et al. Improved monomeric red, orange and yellow fluorescent proteins derived from Discosoma sp. red fluorescent protein. Nat. Biotechnol. 22, 1567–1572 (2004).
Sinha, S., van Nimwegen, E. & Siggia, E.D. A probabilistic method to detect regulatory modules. Bioinformatics 19, i292–i301 (2003).
Di Talia, S., Skotheim, J.M., Bean, J.M., Siggia, E.D. & Cross, F.R. The effects of molecular noise and size control on variability in the budding yeast cell cycle. Nature 448, 947–951 (2007).
Mavrich, T.N. et al. A barrier nucleosome model for statistical positioning of nucleosomes throughout the yeast genome. Genome Res. 18, 1073–1083 (2008).
Crank, J. The Mathematics of Diffusion (Clarendon Press, Oxford, 1986).
Acknowledgements
We thank J.L. Garcia-Cordero for help with modeling phosphate uptake and the Shore lab at the University of Geneva for their assistance with the library construction. This work was supported by a SystemsX.ch grant DynamiX-RTD (2008/005) to S.J.M. and the École Polytechnique Fédérale de Lausanne.
Author information
Authors and Affiliations
Contributions
S.J.M. and A.S.R. designed the library constructs. A.S.R. synthesized the promoter libraries, performed nucleosome occupancy measurements and characterized the promoter library in bulk. N.D. carried out the on-chip experiments. N.D. and S.J.M. designed and implemented the transcription model. A.S.R. and S.J.M. analyzed the data. A.S.R. and S.J.M. wrote the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Figures and Text
Supplementary Figures 1–18 (PDF 10181 kb)
Supplementary data set
Supplementary data set (XLS 607 kb)
Rights and permissions
About this article
Cite this article
Rajkumar, A., Dénervaud, N. & Maerkl, S. Mapping the fine structure of a eukaryotic promoter input-output function. Nat Genet 45, 1207–1215 (2013). https://doi.org/10.1038/ng.2729
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ng.2729