Abstract
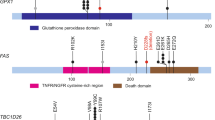
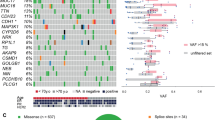
Endometrial cancer is the sixth most commonly diagnosed cancer in women worldwide, causing ∼74,000 deaths annually1. Serous endometrial cancers are a clinically aggressive subtype with a poorly defined genetic etiology2,3,4. We used whole-exome sequencing to comprehensively search for somatic mutations within ∼22,000 protein-encoding genes in 13 primary serous endometrial tumors. We subsequently resequenced 18 genes, which were mutated in more than 1 tumor and/or were components of an enriched functional grouping, from 40 additional serous tumors. We identified high frequencies of somatic mutations in CHD4 (17%), EP300 (8%), ARID1A (6%), TSPYL2 (6%), FBXW7 (29%), SPOP (8%), MAP3K4 (6%) and ABCC9 (6%). Overall, 36.5% of serous tumors had a mutated chromatin-remodeling gene, and 35% had a mutated ubiquitin ligase complex gene, implicating frequent mutational disruption of these processes in the molecular pathogenesis of one of the deadliest forms of endometrial cancer.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
References
Ferlay, J. et al. Estimates of worldwide burden of cancer in 2008: GLOBOCAN 2008. Int. J. Cancer 127, 2893–2917 (2010).
Sherman, M.E. Theories of endometrial carcinogenesis: a multidisciplinary approach. Mod. Pathol. 13, 295–308 (2000).
Hamilton, C.A. et al. Uterine papillary serous and clear cell carcinomas predict for poorer survival compared to grade 3 endometrioid corpus cancers. Br. J. Cancer 94, 642–646 (2006).
Hendrickson, M., Ross, J., Eifel, P., Martinez, A. & Kempson, R. Uterine papillary serous carcinoma: a highly malignant form of endometrial adenocarcinoma. Am. J. Surg. Pathol. 6, 93–108 (1982).
McConechy, M.K. et al. Subtype-specific mutation of PPP2R1A in endometrial and ovarian carcinomas. J. Pathol. 223, 567–573 (2011).
Rudd, M.L. et al. A unique spectrum of somatic PIK3CA (p110α) mutations within primary endometrial carcinomas. Clin. Cancer Res. 17, 1331–1340 (2011).
Shih, I.M. et al. Somatic mutations of PPP2R1A in ovarian and uterine carcinomas. Am. J. Pathol. 178, 1442–1447 (2011).
Hayes, M.P., Douglas, W. & Ellenson, L.H. Molecular alterations of EGFR and PIK3CA in uterine serous carcinoma. Gynecol. Oncol. 113, 370–373 (2009).
National Cancer Institute, SEER Program. SEER Survival Monograph: Cancer Survival Among Adults: U.S. SEER Program, 1988–2001. Patient and Tumor Characteristics (eds. Ries, L.A.G., Young, J.L., Keel, G.E., Eisner, M.P., Lin, Y.D. & Horner, M.-J.) (National Cancer Institute, SEER Program, Bethesda, MD, 2007).
Zhang, Y., LeRoy, G., Seelig, H.P., Lane, W.S. & Reinberg, D. The dermatomyositis-specific autoantigen Mi2 is a component of a complex containing histone deacetylase and nucleosome remodeling activities. Cell 95, 279–289 (1998).
Li, J., Lin, Q., Wang, W., Wade, P. & Wong, J. Specific targeting and constitutive association of histone deacetylase complexes during transcriptional repression. Genes Dev. 16, 687–692 (2002).
Tong, J.K., Hassig, C.A., Schnitzler, G.R., Kingston, R.E. & Schreiber, S.L. Chromatin deacetylation by an ATP-dependent nucleosome remodelling complex. Nature 395, 917–921 (1998).
Xue, Y. et al. NURD, a novel complex with both ATP-dependent chromatin-remodeling and histone deacetylase activities. Mol. Cell 2, 851–861 (1998).
Hall, J.A. & Georgel, P.T. CHD proteins: a diverse family with strong ties. Biochem. Cell Biol. 85, 463–476 (2007).
Lai, A.Y. & Wade, P.A. Cancer biology and NuRD: a multifaceted chromatin remodelling complex. Nat. Rev. Cancer 11, 588–596 (2011).
Polo, S.E., Kaidi, A., Baskcomb, L., Galanty, Y. & Jackson, S.P. Regulation of DNA-damage responses and cell-cycle progression by the chromatin remodelling factor CHD4. EMBO J. 29, 3130–3139 (2010).
Smeenk, G. et al. The NuRD chromatin-remodeling complex regulates signaling and repair of DNA damage. J. Cell Biol. 190, 741–749 (2010).
Chou, D.M. et al. A chromatin localization screen reveals poly (ADP ribose)–regulated recruitment of the repressive polycomb and NuRD complexes to sites of DNA damage. Proc. Natl. Acad. Sci. USA 107, 18475–18480 (2010).
Larsen, D.H. et al. The chromatin-remodeling factor CHD4 coordinates signaling and repair after DNA damage. J. Cell Biol. 190, 731–740 (2010).
Boerkoel, C.F. et al. Mutant chromatin remodeling protein SMARCAL1 causes Schimke immuno-osseous dysplasia. Nat. Genet. 30, 215–220 (2002).
Tsurusaki, Y. et al. Mutations affecting components of the SWI/SNF complex cause Coffin-Siris syndrome. Nat. Genet. 44, 376–378 (2012).
Van Houdt, J.K. et al. Heterozygous missense mutations in SMARCA2 cause Nicolaides-Baraitser syndrome. Nat. Genet. 44, 445–449 (2012).
Welcker, M. & Clurman, B.E. FBW7 ubiquitin ligase: a tumour suppressor at the crossroads of cell division, growth and differentiation. Nat. Rev. Cancer 8, 83–93 (2008).
Cassia, R. et al. Cyclin E gene (CCNE) amplification and hCDC4 mutations in endometrial carcinoma. J. Pathol. 201, 589–595 (2003).
Dutt, A. et al. Drug-sensitive FGFR2 mutations in endometrial carcinoma. Proc. Natl. Acad. Sci. USA 105, 8713–8717 (2008).
Spruck, C.H. et al. hCDC4 gene mutations in endometrial cancer. Cancer Res. 62, 4535–4539 (2002).
Suehiro, Y. et al. Aneuploidy predicts outcome in patients with endometrial carcinoma and is related to lack of CDH13 hypermethylation. Clin. Cancer Res. 14, 3354–3361 (2008).
Forbes, S.A. et al. The Catalogue of Somatic Mutations in Cancer (COSMIC). Curr. Protoc. Hum. Genet. Ch. 10, Unit 10.11 (2008).
Reva, B., Antipin, Y. & Sander, C. Predicting the functional impact of protein mutations: application to cancer genomics. Nucleic Acids Res. 39, e118 (2011).
Wertz, I.E. et al. Sensitivity to antitubulin chemotherapeutics is regulated by MCL1 and FBW7. Nature 471, 110–114 (2011).
Garnett, M.J. et al. Systematic identification of genomic markers of drug sensitivity in cancer cells. Nature 483, 570–575 (2012).
Barbieri, C.E. et al. Exome sequencing identifies recurrent SPOP, FOXA1 and MED12 mutations in prostate cancer. Nat. Genet. 44, 685–689 (2012).
Li, C. et al. Tumor-suppressor role for the SPOP ubiquitin ligase in signal-dependent proteolysis of the oncogenic co-activator SRC-3/AIB1. Oncogene 30, 4350–4364 (2011).
Glaeser, M., Floetotto, T., Hanstein, B., Beckmann, M.W. & Niederacher, D. Gene amplification and expression of the steroid receptor coactivator SRC3 (AIB1) in sporadic breast and endometrial carcinomas. Horm. Metab. Res. 33, 121–126 (2001).
Huang, D.W., Sherman, B.T. & Lempicki, R.A. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 4, 44–57 (2009).
Huang, D.W., Sherman, B.T. & Lempicki, R.A. Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 37, 1–13 (2009).
Dalgliesh, G.L. et al. Systematic sequencing of renal carcinoma reveals inactivation of histone modifying genes. Nature 463, 360–363 (2010).
Fujimoto, A. et al. Whole-genome sequencing of liver cancers identifies etiological influences on mutation patterns and recurrent mutations in chromatin regulators. Nat. Genet. 44, 760–764 (2012).
Grasso, C.S. et al. The mutational landscape of lethal castration-resistant prostate cancer. Nature 487, 239–243 (2012).
Gui, Y. et al. Frequent mutations of chromatin remodeling genes in transitional cell carcinoma of the bladder. Nat. Genet. 43, 875–878 (2011).
Guichard, C. et al. Integrated analysis of somatic mutations and focal copy-number changes identifies key genes and pathways in hepatocellular carcinoma. Nat. Genet. 44, 694–698 (2012).
Jiao, Y. et al. DAXX/ATRX, MEN1, and mTOR pathway genes are frequently altered in pancreatic neuroendocrine tumors. Science 331, 1199–1203 (2011).
Jones, S. et al. Frequent mutations of chromatin remodeling gene ARID1A in ovarian clear cell carcinoma. Science 330, 228–231 (2010).
Ong, C.K. et al. Exome sequencing of liver fluke–associated cholangiocarcinoma. Nat. Genet. 44, 690–693 (2012).
Parsons, D.W. et al. The genetic landscape of the childhood cancer medulloblastoma. Science 331, 435–439 (2011).
Schwartzentruber, J. et al. Driver mutations in histone H3.3 and chromatin remodelling genes in paediatric glioblastoma. Nature 482, 226–231 (2012).
Shain, A.H. et al. Convergent structural alterations define SWItch/Sucrose NonFermentable (SWI/SNF) chromatin remodeler as a central tumor suppressive complex in pancreatic cancer. Proc. Natl. Acad. Sci. USA 109, E252–E259 (2012).
Varela, I. et al. Exome sequencing identifies frequent mutation of the SWI/SNF complex gene PBRM1 in renal carcinoma. Nature 469, 539–542 (2011).
Wiegand, K.C. et al. ARID1A mutations in endometriosis-associated ovarian carcinomas. N. Engl. J. Med. 363, 1532–1543 (2010).
Wang, K. et al. Exome sequencing identifies frequent mutation of ARID1A in molecular subtypes of gastric cancer. Nat. Genet. 43, 1219–1223 (2011).
Ogryzko, V.V., Schiltz, R.L., Russanova, V., Howard, B.H. & Nakatani, Y. The transcriptional coactivators p300 and CBP are histone acetyltransferases. Cell 87, 953–959 (1996).
Huang, J., Zhao, Y.L., Li, Y., Fletcher, J.A. & Xiao, S. Genomic and functional evidence for an ARID1A tumor suppressor role. Genes Chromosom. Cancer 46, 745–750 (2007).
Guan, B., Wang, T.L. & Shih, I.M. ARID1A, a factor that promotes formation of SWI/SNF-mediated chromatin remodeling, is a tumor suppressor in gynecologic cancers. Cancer Res. 71, 6718–6727 (2011).
Hargreaves, D.C. & Crabtree, G.R. ATP-dependent chromatin remodeling: genetics, genomics and mechanisms. Cell Res. 21, 396–420 (2011).
Guan, B. et al. Mutation and loss of expression of ARID1A in uterine low-grade endometrioid carcinoma. Am. J. Surg. Pathol. 35, 625–632 (2011).
McConechy, M.K. et al. Use of mutation profiles to refine the classification of endometrial carcinomas. J. Pathol. 228, 20–30 (2012).
Wiegand, K.C. et al. Loss of BAF250a (ARID1A) is frequent in high-grade endometrial carcinomas. J. Pathol. 224, 328–333 (2011).
Urick, M.E. et al. PIK3R1 (p85α) is somatically mutated at high frequency in primary endometrial cancer. Cancer Res. 71, 4061–4067 (2011).
Kuhn, E. et al. Identification of molecular pathway aberrations in uterine serous carcinoma by genome-wide analyses. J. Natl. Cancer Inst. 104, 1503–1513 (2012).
Teer, J.K. et al. Systematic comparison of three genomic enrichment methods for massively parallel DNA sequencing. Genome Res. 20, 1420–1431 (2010).
Teer, J.K., Green, E.D., Mullikin, J.C. & Biesecker, L.G. VarSifter: visualizing and analyzing exome-scale sequence variation data on a desktop computer. Bioinformatics 28, 599–600 (2012).
Benjamini, Y. & Hochberg, Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J.R. Stat. Soc. 57, 289–300 (1995).
Sjöblom, T. et al. The consensus coding sequences of human breast and colorectal cancers. Science 314, 268–274 (2006).
Rubin, A.F. & Green, P. Comment on “The consensus coding sequences of human breast and colorectal cancers”. Science 317, 1500 (2007).
Acknowledgements
We thank our colleagues for critical reading of the manuscript; R.T. Moreland and N. Trivedi of the National Human Genome Research Institute Bioinformatics and Scientific Programming Core, respectively, for performing in silico PCR and giving advice on statistics; and J. Teer for sharing expertise on VarSifter. A. Santin (Yale School of Medicine) kindly provided the ARK1 and ARK2 cell lines. The study was funded in part by the Intramural Program of the National Human Genome Research Institute, US NIH (D.W.B., J.C.M. and M.J.M.); NIH grant R01CA112021 (D.C.S.); the Avon Foundation (D.C.S.); NIH grant R01CA140323 (A.K.G.); and the Ovarian Cancer Fund (A.K.G.). P.H. is supported by grants from the NIH (CA016519) and by the Canadian Institutes for Health Research (MOP-38096).
Author information
Authors and Affiliations
Consortia
Contributions
D.W.B. designed and directed the study and wrote the manuscript. A.K.G. contributed clinical specimens. M.J.M. and D.C.S. conducted pathological review of clinical specimens. M.L.R. prepared DNA samples and performed identity testing and microsatellite instability analysis. NISC performed library construction and whole-exome sequencing. NISC and N.F.H. performed variant calling. M.L.G. and A.J.O. curated and orthogonally validated exome sequencing data. M.L.G., A.J.O. and D.W.B. interpreted the exome data and established filtering criteria. M.L.G., A.J.O., M.L.R., J.C.P., B.M.E., S.Z. and D.W.B. designed, performed, analyzed and interpreted the mutation prevalence screens. A.J.O. and M.L.G. analyzed MSH6. M.E.U. and M.L.G. generated sequence conservation alignments. M.E.U. performed cell culture and immunoblotting. N.F.H., M.L.G. and J.C.M. performed statistical analyses. N.F.H. performed the power calculation. D.W.B., M.E.U., M.L.G., M.L.R., A.J.O., N.F.H., J.C.M., A.K.G., P.H. and N.J.O. edited and commented on the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Additional information
A list of members appears in the Supplementary Note.
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–10, Supplementary Tables 2, 3, 6–10, 12, 14 and 15 and Supplementary Note (PDF 2801 kb)
Supplementary Table 1
Characteristics of endometrial tumors included in the discovery and prevalence screens (XLS 34 kb)
Supplementary Table 4
Filtered exonic and splice junction somatic mutations in a hypermutated tumor (T155) in the discovery screen (XLS 506 kb)
Supplementary Table 5
Filtered exonic and splice junction somatic mutations among 12 tumors in the discovery screen (XLS 272 kb)
Supplementary Table 11
Microsatellite instability (MSI) status and MSH6 status of 160 tumors included in the discovery and prevalence screens of CHD4, FBXW7, and SPOP (XLS 29 kb)
Supplementary Table 13
Enriched functional groupings identified by DAVID analysis (XLS 285 kb)
Supplementary Table 16
All exonic and splice junction somatic mutations among 12 tumors in the discovery screen (XLS 396 kb)
Supplementary Table 17
All exonic and splice junction somatic mutations in a hypermutated tumor (T155) in the discovery screen (XLS 518 kb)
Supplementary Table 18
Primers used for PCR amplification (XLS 219 kb)
Rights and permissions
About this article
Cite this article
Le Gallo, M., O'Hara, A., Rudd, M. et al. Exome sequencing of serous endometrial tumors identifies recurrent somatic mutations in chromatin-remodeling and ubiquitin ligase complex genes. Nat Genet 44, 1310–1315 (2012). https://doi.org/10.1038/ng.2455
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ng.2455
This article is cited by
-
The epigenetic factor CHD4 contributes to metastasis by regulating the EZH2/β-catenin axis and acts as a therapeutic target in ovarian cancer
Journal of Translational Medicine (2023)
-
Experimental analysis of bladder cancer-associated mutations in EP300 identifies EP300-R1627W as a driver mutation
Molecular Medicine (2023)
-
SPOP mutations promote tumor immune escape in endometrial cancer via the IRF1–PD-L1 axis
Cell Death & Differentiation (2023)
-
Sex specific regulation of TSPY-Like 2 in the DNA damage response of cancer cells
Cell Death & Disease (2023)
-
Loss-of-function mutations of SOX17 lead to YAP/TEAD activation-dependent malignant transformation in endometrial cancer
Oncogene (2023)