Abstract
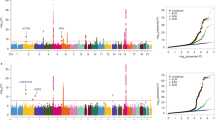
To identify common genetic variants that contribute to lung cancer susceptibility, we conducted a multistage genome-wide association study of lung cancer in Asian women who never smoked. We scanned 5,510 never-smoking female lung cancer cases and 4,544 controls drawn from 14 studies from mainland China, South Korea, Japan, Singapore, Taiwan and Hong Kong. We genotyped the most promising variants (associated at P < 5 × 10−6) in an additional 1,099 cases and 2,913 controls. We identified three new susceptibility loci at 10q25.2 (rs7086803, P = 3.54 × 10−18), 6q22.2 (rs9387478, P = 4.14 × 10−10) and 6p21.32 (rs2395185, P = 9.51 × 10−9). We also confirmed associations reported for loci at 5p15.33 and 3q28 and a recently reported finding at 17q24.3. We observed no evidence of association for lung cancer at 15q25 in never-smoking women in Asia, providing strong evidence that this locus is not associated with lung cancer independent of smoking.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout


Similar content being viewed by others
References
Sun, S., Schiller, J.H. & Gazdar, A.F. Lung cancer in never smokers—a different disease. Nat. Rev. Cancer 7, 778–790 (2007).
Sun, Y. et al. Lung adenocarcinoma from East Asian never-smokers is a disease largely defined by targetable oncogenic mutant kinases. J. Clin. Oncol. 28, 4616–4620 (2010).
Rudin, C.M. et al. Lung cancer in never smokers: molecular profiles and therapeutic implications. Clin. Cancer Res. 15, 5646–5661 (2009).
Thun, M.J. et al. Lung cancer occurrence in never-smokers: an analysis of 13 cohorts and 22 cancer registry studies. PLoS Med. 5, e185 (2008).
Gao, Y.T. et al. Lung cancer among Chinese women. Int. J. Cancer 40, 604–609 (1987).
Gu, D. et al. Cigarette smoking and exposure to environmental tobacco smoke in China: the international collaborative study of cardiovascular disease in Asia. Am. J. Public Health 94, 1972–1976 (2004).
Lan, Q., Chapman, R.S., Schreinemachers, D.M., Tian, L. & He, X. Household stove improvement and risk of lung cancer in Xuanwei, China. J. Natl. Cancer Inst. 94, 826–835 (2002).
Couraud, S., Zalcman, G., Milleron, B., Morin, F. & Souquet, P.J. Lung cancer in never smokers—a review. Eur. J. Cancer 48, 1299–1311 (2012).
Samet, J.M. et al. Lung cancer in never smokers: clinical epidemiology and environmental risk factors. Clin. Cancer Res. 15, 5626–5645 (2009).
Lo, Y.L. et al. Risk factors for primary lung cancer among never smokers by gender in a matched case-control study. Cancer Causes Control published online, doi:10.1007/s10552-012-9994-x (22 May 2012).
Hsiung, C.A. et al. The 5p15.33 locus is associated with risk of lung adenocarcinoma in never-smoking females in Asia. PLoS Genet. 6, e1001051 (2010).
Landi, M.T. et al. A genome-wide association study of lung cancer identifies a region of chromosome 5p15 associated with risk for adenocarcinoma. Am. J. Hum. Genet. 85, 679–691 (2009).
Hosgood, H.D. III et al. Genetic variant in TP63 on locus 3q28 is associated with risk of lung adenocarcinoma among never-smoking females in Asia. Hum. Genet. 131, 1197–1203 (2012).
Miki, D. et al. Variation in TP63 is associated with lung adenocarcinoma susceptibility in Japanese and Korean populations. Nat. Genet. 42, 893–896 (2010).
Amundadottir, L. et al. Genome-wide association study identifies variants in the ABO locus associated with susceptibility to pancreatic cancer. Nat. Genet. 41, 986–990 (2009).
de Bakker, P.I. et al. Practical aspects of imputation-driven meta-analysis of genome-wide association studies. Hum. Mol. Genet. 17, R122–R128 (2008).
Hu, Z. et al. A genome-wide association study identifies two new lung cancer susceptibility loci at 13q12.12 and 22q12.2 in Han Chinese. Nat. Genet. 43, 792–796 (2011).
McKay, J.D. et al. Lung cancer susceptibility locus at 5p15.33. Nat. Genet. 40, 1404–1406 (2008).
Truong, T. et al. Replication of lung cancer susceptibility loci at chromosomes 15q25, 5p15, and 6p21: a pooled analysis from the International Lung Cancer Consortium. J. Natl. Cancer Inst. 102, 959–971 (2010).
Shiraishi, K. et al. A genome-wide association study identifies two new susceptibility loci for lung adenocarcinoma in the Japanese population. Nat. Genet. 44, 900–903 (2012).
Amos, C.I. et al. Genome-wide association scan of tag SNPs identifies a susceptibility locus for lung cancer at 15q25.1. Nat. Genet. 40, 616–622 (2008).
Hung, R.J. et al. A susceptibility locus for lung cancer maps to nicotinic acetylcholine receptor subunit genes on 15q25. Nature 452, 633–637 (2008).
Wang, Y. et al. Common 5p15.33 and 6p21.33 variants influence lung cancer risk. Nat. Genet. 40, 1407–1409 (2008).
Wu, C. et al. Genetic variants on chromosome 15q25 associated with lung cancer risk in Chinese populations. Cancer Res. 69, 5065–5072 (2009).
Shi, J. et al. Inherited variation at chromosome 12p13.33, including RAD52, influences the risk of squamous cell lung carcinoma. Cancer Discov. 2, 131–139 (2012).
Dong, J. et al. Association analyses identify multiple new lung cancer susceptibility loci and their interactions with smoking in the Chinese population. Nat. Genet. 44, 895–899 (2012).
Wang, Z. et al. Improved imputation of common and uncommon SNPs with a new reference set. Nat. Genet. 44, 6–7 (2012).
Howie, B.N., Donnelly, P. & Marchini, J. A flexible and accurate genotype imputation method for the next generation of genome-wide association studies. PLoS Genet. 5, e1000529 (2009).
Ramirez, D.M., Khvotchev, M., Trauterman, B. & Kavalali, E.T. Vti1a identifies a vesicle pool that preferentially recycles at rest and maintains spontaneous neurotransmission. Neuron 73, 121–134 (2012).
Kunwar, A.J. et al. Lack of the endosomal SNAREs vti1a and vti1b led to significant impairments in neuronal development. Proc. Natl. Acad. Sci. USA 108, 2575–2580 (2011).
Bose, A. et al. The v-SNARE Vti1a regulates insulin-stimulated glucose transport and Acrp30 secretion in 3T3-L1 adipocytes. J. Biol. Chem. 280, 36946–36951 (2005).
Petridou, E.T. et al. Circulating adiponectin levels and expression of adiponectin receptors in relation to lung cancer: two case-control studies. Oncology 73, 261–269 (2007).
Bass, A.J. et al. Genomic sequencing of colorectal adenocarcinomas identifies a recurrent VTI1A-TCF7L2 fusion. Nat. Genet. 43, 964–968 (2011).
Lemmon, M.A. & Schlessinger, J. Cell signaling by receptor tyrosine kinases. Cell 141, 1117–1134 (2010).
Acquaviva, J., Wong, R. & Charest, A. The multifaceted roles of the receptor tyrosine kinase ROS in development and cancer. Biochim. Biophys. Acta 1795, 37–52 (2009).
Li, C. et al. Spectrum of oncogenic driver mutations in lung adenocarcinomas from East Asian never smokers. PLoS ONE 6, e28204 (2011).
Rikova, K. et al. Global survey of phosphotyrosine signaling identifies oncogenic kinases in lung cancer. Cell 131, 1190–1203 (2007).
Takeuchi, K. et al. RET, ROS1 and ALK fusions in lung cancer. Nat. Med. 18, 378–381 (2012).
Bergethon, K. et al. ROS1 rearrangements define a unique molecular class of lung cancers. J. Clin. Oncol. 30, 863–870 (2012).
Kim, M. et al. Epigenetic down-regulation and suppressive role of DCBLD2 in gastric cancer cell proliferation and invasion. Mol. Cancer Res. 6, 222–230 (2008).
Koshikawa, K. et al. Significant up-regulation of a novel gene, CLCP1, in a highly metastatic lung cancer subline as well as in lung cancers in vivo. Oncogene 21, 2822–2828 (2002).
Nagai, H. et al. CLCP1 interacts with semaphorin 4B and regulates motility of lung cancer cells. Oncogene 26, 4025–4031 (2007).
Silverberg, M.S. et al. Ulcerative colitis–risk loci on chromosomes 1p36 and 12q15 found by genome-wide association study. Nat. Genet. 41, 216–220 (2009).
Urayama, K.Y. et al. Genome-wide association study of classical Hodgkin lymphoma and Epstein-Barr virus status–defined subgroups. J. Natl. Cancer Inst. 104, 240–253 (2012).
Nakanishi, K. & Shima, Y. Capture of type 1 diabetes–susceptible HLA DR-DQ haplotypes in Japanese subjects using a tag single nucleotide polymorphism. Diabetes Care 33, 162–164 (2010).
Chanock, S.J. & Hunter, D.J. Genomics: when the smoke clears. Nature 452, 537–538 (2008).
Spitz, M.R., Amos, C.I., Dong, Q., Lin, J. & Wu, X. The CHRNA5-A3 region on chromosome 15q24-25.1 is a risk factor both for nicotine dependence and for lung cancer. J. Natl. Cancer Inst. 100, 1552–1556 (2008).
Wang, Y., Broderick, P., Matakidou, A., Eisen, T. & Houlston, R.S. Chromosome 15q25 (CHRNA3-CHRNA5) variation impacts indirectly on lung cancer risk. PLoS ONE 6, e19085 (2011).
Patterson, N., Price, A.L. & Reich, D. Population structure and eigenanalysis. PLoS Genet. 2, e190 (2006).
Price, A.L. et al. Principal components analysis corrects for stratification in genome-wide association studies. Nat. Genet. 38, 904–909 (2006).
Fearnhead, P. SequenceLDhot: detecting recombination hotspots. Bioinformatics 22, 3061–3066 (2006).
Fearnhead, P., Harding, R.M., Schneider, J.A., Myers, S. & Donnelly, P. Application of coalescent methods to reveal fine-scale rate variation and recombination hotspots. Genetics 167, 2067–2081 (2004).
Abnet, C.C. et al. Genotypic variants at 2q33 and risk of esophageal squamous cell carcinoma in China: a meta-analysis of genome-wide association studies. Hum. Mol. Genet. 21, 2132–2141 (2012).
Luna, A. & Nicodemus, K.K. snp.plotter: an R-based SNP/haplotype association and linkage disequilibrium plotting package. Bioinformatics 23, 774–776 (2007).
Acknowledgements
We thank J.-J. Yang, X.-N. Yang, Q. Zhou, W.-B. Guo, S.-L. Chen, Y. Huang, Z. Xie, J.-G. Chen, H.-H. Yan, K. Tajima, Y. Yatabe, T. Hida, K.-L. Chuah, A. Ng, P. Eng, S.-S. Leong, M.-K. Ang, E. Lim, T.-K. Lim, M. Teh, W.-T. Poh and A. Tee. The overall GWAS project was supported by the intramural program of the US National Institutes of Health/National Cancer Institute. A list of support provided to individual studies is provided in the Supplementary Note.
Author information
Authors and Affiliations
Contributions
Q.L., N.R., S.J.C., D. Lin, C.A.H., Y.-C.H., K.M., A.S., H.D.H., J.Y.P., C.-J.C., Y.H.K., Y.T.K., C.L., Y.-L.W., P.-C.Y., B.Z., M.-H.S., J.F.F., K.C., W.Z., T.W., H.S., I.-S.C., D. Lu, N. Caporaso, W.P., R.K., J. Liu, M.T.L., N. Chatterjee, M.T. and M.Y. organized and designed the study. S.J.C., D. Lin, R.K., J. Liu, C.A.H., K.M., T.W., L.B., M.Y., J. Yuenger, Z.Y., C.W., H.G., A.H., W.W., Y.L., W.P., H.-C.L. and B.Z. conducted and supervised the genotyping of samples. Z.W., K.B.J., N.R., Q.L., S.J.C., N. Chatterjee, C.A.H., H.D.H., W.H., M.Y., I.-S.C., C.-F.H., W.-C.W., C.C.C., S.I.B., C.-H.C., R.V. and Y.-H.C. contributed to the design and execution of statistical analysis. Q.L., N.R., S.J.C., Z.W., W.H., C.C.C., C.A.H., K.M., Y.-C.H., A.S., H.D.H., N. Chatterjee, N. Caporaso, C.L., M.Y., B.A.B., M.T., S.-J.A., S.I.B., M.T.L., C.K., R.V., Y.-L.W., J.F.F. and I.-S.C. wrote the first draft of the manuscript. C.A.H., Q.L., B.Z., Y.-C.H., K.M., A.S., K.C., J.-C.W., M.P.W., W.Z., J.Y.P., W.H., C.-J.C., Y.H.K., Y.T.K., T.W., H.S., I.-S.C., T.M., H.N.K., F.W., Z.Y., C.W., S.-J.A., G.-C.C., B.Q., V.H.F.L., D. Lu, H.-S.J., J.S.S., J.H.K., Y.-T.G., Y.-H.T., Y.J.J., H.G., Z.H., I.-J.O., K.-Y.C., X.H., W.W., J.C., X.-C.Z., M.-S.H., H.Z., J. Wang, X.Z., J.E.C., W.-C.S., K.H.P., S.W.S., X.-O.S., Y.-M.C., L.L., C.H.K., L.H., Y.-C.K., T.-Y.Y., J.X., P.G., W.T., J.S., C.-L.W., H.L., A.D.L.S., Z.Z., Y.C., Y.Y.C., J.-Y.H., J.S.K., H.-I.Y., Q.C., C.-C.L., I.K.P., P.X., J.D., Q.H., R.-P.P., T.K., S.-S.K., C.-Y.C., R.V., J. Wu, W.-Y.L., K.-C.C., W.-H.C., B.-T.J., J.K.C.C., M.C., Y.-J.L., J. Yokota, J. Li, H.C., Y.-B.X., C.-J.Y., H.K., G.W., L.J., Y.-L.L., K.S., Y.-L.W., P.-C.Y., M.-H.S., J.F.F., D. Lin, S.J.C. and N.R. conducted the epidemiological studies and contributed samples to the GWAS and/or follow-up genotyping. All authors contributed to the writing of the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Text and Figures
Supplementary Note, Supplementary Tables 1–6 and Supplementary Figures 1–5 (PDF 2104 kb)
Rights and permissions
About this article
Cite this article
Lan, Q., Hsiung, C., Matsuo, K. et al. Genome-wide association analysis identifies new lung cancer susceptibility loci in never-smoking women in Asia. Nat Genet 44, 1330–1335 (2012). https://doi.org/10.1038/ng.2456
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ng.2456
This article is cited by
-
E-GWAS: an ensemble-like GWAS strategy that provides effective control over false positive rates without decreasing true positives
Genetics Selection Evolution (2023)
-
Sequre: a high-performance framework for secure multiparty computation enables biomedical data sharing
Genome Biology (2023)
-
Contribution of an Asian-prevalent HLA haplotype to the risk of HBV-related hepatocellular carcinoma
Scientific Reports (2023)
-
Ethnicity-specific association between TERT rs2736100 (A > C) polymorphism and lung cancer risk: a comprehensive meta-analysis
Scientific Reports (2023)
-
Lung cancer in never smokers (LCINS): development of a UK national research strategy
BJC Reports (2023)