Abstract
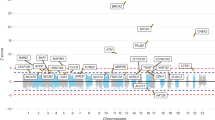
We conducted a genome-wide association study of male breast cancer comprising 823 cases and 2,795 controls of European ancestry, with validation in independent sample sets totaling 438 cases and 474 controls. A SNP in RAD51B at 14q24.1 was significantly associated with male breast cancer risk (P = 3.02 × 10−13; odds ratio (OR) = 1.57). We also refine association at 16q12.1 to a SNP within TOX3 (P = 3.87 × 10−15; OR = 1.50).
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout

Similar content being viewed by others
References
Bevier, M., Sundquist, K. & Hemminki, K. Breast Cancer Res. Treat. 132, 723–728 (2012).
Basham, V.M. et al. Breast Cancer Res. 4, R2 (2002).
Thomas, G. et al. Nat. Genet. 41, 579–584 (2009).
Wang, C. et al. Mol. Endocrinol. 25, 1527–1538 (2011).
Orr, N. et al. PLoS Genet. 7, e1002290 (2011).
Meijers-Heijboer, H. et al. Nat. Genet. 31, 55–59 (2002).
Easton, D.F. et al. Nature 447, 1087–1093 (2007).
Haiman, C.A. et al. Nat. Genet. 43, 1210–1214 (2011).
Stevens, K.N. et al. Cancer Res. 72, 1795–1803 (2012).
Fletcher, O. et al. J. Natl. Cancer Inst. 103, 425–435 (2011).
Stacey, S.N. et al. Nat. Genet. 39, 865–869 (2007).
Stacey, S.N. et al. Nat. Genet. 40, 703–706 (2008).
Turnbull, C. et al. Nat. Genet. 42, 504–507 (2010).
Ghoussaini, M. et al. Nat. Genet. 44, 312–318 (2012).
Garcia-Closas, M. et al. PLoS Genet. 4, e1000054 (2008).
Acknowledgements
We thank the men who participated in the study. For ICR sample collection, we are grateful to B. Smith, D. Hogben, R. McCann, J. Melia, S. Squires, M. Snigorska, J. Micic and K. Sibley for administrative help and advice; to the research nurses A. Butlin, M. Pelerin, J. Wood and T. Gardener for collection of blood samples and questionnaire data from participants; to D. Thomas and her team from the Breakthrough Generations Study for coordinating collection of controls; and to the cancer registries of England and Wales for providing information on eligible participants. The Study of Epidemiology and Risk Factors in Cancer Heredity (SEARCH) thanks the SEARCH team. We wish to thank H. Thorne, E. Niedermayr, all the kConFab research nurses and staff, the heads and staff of the Family Cancer Clinics and the Clinical Follow Up Study (funded from 2001–2009 by the Australian National Health & Medical Research Council (NHMRC) and currently by the National Breast Cancer Foundation and Cancer Australia, 628333) for their contributions to this resource and the many families who contribute to kConFab. We thank G. Wilhoite for preparing the US samples for this study. We thank A. Jukkola-Vuorinen, V. Kataja and P. Auvinen, as well as S. Hallamies and S. Ranta, for their contributions to the Finnish Male Breast Cancer Study. We thank J. Forteza, M. Fraga, C. Celeiro and the staff of the Department of Pathology of CHUS Santiago and A. Carracedo and C.M. Redondo for their contributions to the Spanish study. Finally, we thank the consultants who cared for the study participants for their advice and help. The Breakthrough Male Breast Cancer Study was approved by the South Eastern Research Ethics Committee. Participants gave informed consent for all aspects of the study.
This work was supported by Breakthrough Breast Cancer, Movember and the Institute of Cancer Research who acknowledge National Health Service (NHS) funding to the National Institute for Health Research (NIHR) Biomedical Research Centre. The funders had no role in study design, data collection and analysis, decision to publish or preparation of the manuscript. The High-Throughput Genomics Group at the Wellcome Trust Centre for Human Genetics is funded by Wellcome Trust grant 090532/Z/09/Z and UK Medical Research Council (MRC) Hub grant G0900747 91070. This work made use of data and samples generated by the 1958 British Birth Cohort under grant G0000934 from the UK MRC and grant 068545/Z/02 from the Wellcome Trust. The Finnish Male Breast Cancer Study acknowledges funding from the Finnish Breast Cancer Group, the Finnish Oncology Association and the Salonoja Foundation. L.O. acknowledges funding from the Italian Association for Cancer Research (AIRC, IG 8713). D.P. acknowledges funding from the Istituto Toscano Tumori, Florence, Italy. SEARCH is funded by a grant from Cancer Research UK (C490/A10124) and is supported by the University of Cambridge NIHR Biomedical Research Centre. A.M.D. has been funded by Cancer Research UK grant CA8197/A10865 and by the Joseph Mitchell Fund. D.T.B. acknowledges funding by Cancer Research UK (Programme Award C588/A10589). M.G.-D. and J.E.C. acknowledge the support of the Department of Pathology of the CHUS Santiago Hospital, the Galician Foundation of Genomic Medicine and the Program Grupos Emergentes, Cancer Genetics Unit of CHUVI, Instituto de Salud Carlos III. H.O. and I.H. acknowledge funding from the Swedish Cancer Society and The Swedish Medical Research Council. The collection of samples and epidemiological data for the US cases was supported by US ARMY Grant DAMD-17-96-I-6266 and US National Institutes of Health (NIH) grant R01CA74415. S.L.N. was partially supported by the Morris and Horowitz Families Endowed Professorship at the City of Hope.
Author information
Authors and Affiliations
Consortia
Contributions
A.A. and A.J.S. conceived the Breakthrough Breast Cancer Male Breast Cancer Study and obtained financial support. N.O. and A.J.S. designed the GWAS. N.O. drafted the manuscript with substantial input from O.F., M.J., C.J.L., R.S.H., M.G.-C., A.A. and A.J.S. N.O., R.C., S.S.M., C.M. and M.Z. performed statistical and bioinformatics analyses. K.T. managed sample handling and DNA extraction. A.L., N.J., G.B. and C.B. performed genotyping. A.H.T. and P.A.J. coordinated sample collection at the Peter MacCallum Cancer Centre; S.E.B. and S.B. coordinated sample collection at Copenhagen University Hospital; H.N. and J.M. coordinated sample collection for the Finnish Male Breast Cancer Study; E.F. and Y.L. coordinate sample collection at Sheba Medical Centre and the Sackler School of Medicine; D.P., G.M. and I.Z. coordinated sample collection at ISPO (Cancer Research and Prevention Institute); L.O. and G.G. coordinated sample collection at Sapienza University of Rome; A.H. and A.M.W.v.d.O. coordinated sample collection at Erasmus University Medical Center; S.N. and M.K. coordinated sample collection at the Institute of Oncology Ljubljana; M.G.-D. and J.E.C. coordinated sample collection in Galicia, Spain; H.O. and I.H. coordinated sample collection at Lund University; D.F.E., P.D.P.P. and A.M.D. coordinated sample collection for the SEARCH/Cambridge studies; D.T.B. coordinated sample collection at the Leeds Institute of Molecular Medicine; and S.L.N. and L.S. coordinated collection of US samples. All authors contributed to the final manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Additional information
A list of members and affiliations is provided in the Supplementary Note.
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–7, Supplementary Tables 1–7, Supplementary Methods and Supplementary Note (PDF 1917 kb)
Rights and permissions
About this article
Cite this article
Orr, N., Lemnrau, A., Cooke, R. et al. Genome-wide association study identifies a common variant in RAD51B associated with male breast cancer risk. Nat Genet 44, 1182–1184 (2012). https://doi.org/10.1038/ng.2417
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ng.2417
This article is cited by
-
Infertility and risk of breast cancer in men: a national case–control study in England and Wales
Breast Cancer Research (2022)
-
A truncating variant of RAD51B associated with primary ovarian insufficiency provides insights into its meiotic and somatic functions
Cell Death & Differentiation (2022)
-
Prevalence of mammary Paget’s disease in urban China in 2016
Scientific Reports (2021)
-
Germline RAD51B variants confer susceptibility to breast and ovarian cancers deficient in homologous recombination
npj Breast Cancer (2021)
-
Male and female breast cancer: the two faces of the same genetic susceptibility coin
Breast Cancer Research and Treatment (2021)