Abstract
A highly invasive form of non-typhoidal Salmonella (iNTS) disease has recently been documented in many countries in sub-Saharan Africa. The most common Salmonella enterica serovar causing this disease is Typhimurium (Salmonella Typhimurium). We applied whole-genome sequence–based phylogenetic methods to define the population structure of sub-Saharan African invasive Salmonella Typhimurium isolates and compared these to global Salmonella Typhimurium populations. Notably, the vast majority of sub-Saharan invasive Salmonella Typhimurium isolates fell within two closely related, highly clustered phylogenetic lineages that we estimate emerged independently ∼52 and ∼35 years ago in close temporal association with the current HIV pandemic. Clonal replacement of isolates from lineage I by those from lineage II was potentially influenced by the use of chloramphenicol for the treatment of iNTS disease. Our analysis suggests that iNTS disease is in part an epidemic in sub-Saharan Africa caused by highly related Salmonella Typhimurium lineages that may have occupied new niches associated with a compromised human population and antibiotic treatment.
Similar content being viewed by others
Main
S. enterica is a diverse bacterial species that remains a common cause of infectious disease in humans and animals throughout the world1. Human Salmonella infections are classically divided into diseases caused by typhoidal Salmonella or non-typhoidal Salmonella (NTS). The former category includes the human-restricted S. enterica serovars Typhi and Paratyphi that cause the systemic disease typhoid, whereas NTS is comprised of the majority of the other serovars that predominantly cause self-limiting gastroenteritis in humans2. S. enterica serovar Typhi (Salmonella Typhi) is a human-restricted pathogen that is transmitted from human to human, whereas NTS disease is normally associated with zoonotic Salmonella reservoirs, typically domesticated animals, with little or no sustained human-to-human transmission.
In contrast to this classical view, NTS are a frequent cause of invasive bacterial disease in many countries in sub-Saharan Africa3,4. This invasive form of NTS disease (iNTS) is common both in children with malnutrition, severe anemia, malaria or HIV4,5 and in HIV-infected adults6, frequently surpassing Salmonella Typhi in many parts of the region as the dominant cause of invasive salmonellosis. The clinical presentation of iNTS disease is distinct from those of both gastroenteritis and typhoid fever and is characterized by a nonspecific fever that can be indistinguishable from malaria and in rare cases is accompanied by diarrhea7. The frequency of NTS-associated case fatalities can be extremely high in both adults and children (22–45%)6,8,9,10.
S. enterica serovar Typhimurium (Salmonella Typhimurium) is one of the serovars that is most frequently associated with iNTS in the sub-Saharan region, although other serovars, including S. enterica serovar Enteritidis, have also been implicated3,4,8. We previously reported that Salmonella Typhimurium isolates from Kenya and Malawi were predominantly of a new multilocus sequence type (MLST) designated ST313 (ref. 7) that is rarely isolated from outside sub-Saharan Africa. The DNA sequence of representative multidrug-resistant (MDR) ST313 isolates D23580 and A130 identified genomic features distinct from those of previously characterized gastroenteritis-associated strains7. These features included evidence of partial genome degradation, with some parallels to that observed in the S. Enterica serovars Typhi and Paratyphi A that has been linked to niche adaptation11,12.
Here, we use SNP-based phylogenetic methods based on whole-genome sequences to determine the population structure of a geographically diverse collection of invasive Salmonella Typhimurium isolates from different sub-Saharan African countries. These data are placed in the phylogenetic context of Salmonella Typhimurium isolates from other parts of the world. We provide evidence that two tightly clustered genetic lineages have emerged within the last 60 years to be the dominant cause of epidemic invasive Salmonella Typhimurium disease in the region. We highlight the potential role of antibiotic resistance acquisition in driving the epidemic and the temporal association of iNTS disease with an increased prevalence of HIV.
Results
Phylogenetic analysis of Salmonella Typhimurium
Salmonella Typhimurium represents an unstratified serologically defined group within the broader species S. enterica13. Therefore, to place the invasive Salmonella Typhimurium isolates from sub-Saharan Africa into an evolutionary and phylogenetic context, we exploited whole-genome sequencing to discover potentially informative SNPs within a collection of 179 Salmonella Typhimurium isolates that were collected between 1938 and 2010 from different parts of the world. Our collection included 129 invasive Salmonella Typhimurium isolates from Malawi, Kenya, Mozambique, Uganda, The Democratic Republic of Congo (DRC), Nigeria and Mali (Supplementary Table 1). Data were available for 10,623 high-quality SNPs, corresponding to approximately 1 SNP for every 407 bp, that were distributed relatively uniformly across the genome of the reference Salmonella Typhimurium SL1344. To refine phylogenetic analysis, SNPs associated with repetitive sequences, mobile elements and phage sequences, representing ∼4% of the genome, were excluded. We detected no evidence of extensive recombination within the remaining genomic sequences, and, consequently, SNPs mapping to these regions were used to reconstruct a maximum-likelihood phylogenetic tree14 (Fig. 1).
Unrooted maximum-likelihood tree showing the relationship between isolates associated with invasive disease and gastroenteritis. Lineages of human invasive Salmonella Typhimurium are shown in red and labeled I–3IV. The phylogenetic positions of invasive strains A130 (ref. 7) and D23580 (ref. 7) and gastroenteritis-associated strains DT104, LT2 and SL1344 are indicated. Branch lengths are indicative of the estimated substitution rate per variable site. Scale bar, 0.009 substitutions per variable site. The numbers of isolates (in parentheses) and MLST groups are indicated in boxes. Top left, unrooted maximum-likelihood tree of plasmid sequences showing congruence with the chromosomal tree. Asterisks indicate nodes with 100% bootstrap support.
Notably, invasive Salmonella Typhimurium isolates from sub-Saharan Africa fall predominantly into two distinct ST313 phylogenetic lineages designated as lineages I and II. Furthermore, these lineages form distinct and extremely tight clusters on separate branches from other Salmonella Typhimurium that were isolated elsewhere in the world. The tight clustering is illustrated by the fact that isolates within either lineage I or lineage II are separated by mean differences of as few as 33 and 21 SNPs, respectively. Isolates in lineage I are distinguished from those of lineage II by an average of 455 SNPs and from other Salmonella Typhimurium isolates by >700 SNPs. Both lineages are thus more closely related to each other than they are to any other Salmonella Typhimurium isolate within the tree. The two invasive Salmonella Typhimurium lineages are joined to the main tree by relatively long branches, but there is divergence at the branch tips, suggesting recent clonal or population expansion. MLST analysis confirmed lineages I and II as ST313, although a single isolate, 5580, from lineage I is ST394, which is a single-locus variant of ST313 (Supplementary Fig. 1). All eight invasive Salmonella Typhimurium isolates from sub-Saharan Africa that fall outside of lineages I and II are ST19, a common sequence type to which 82% (41/50) of the non-African Salmonella Typhimurium isolates that we sequenced belong. Other sequence types represented in the non–sub-Saharan Salmonella Typhimurium lineages include ST34 (5/50), ST98 (1/50), ST128 (2/50) and ST568 (2/50) (Supplementary Fig. 1).
Temporal and geographic distribution relative to phylogeny
We performed BEAST15 analysis on 129 sub-Saharan invasive Salmonella Typhimurium isolates from 7 sub-Saharan African countries covering a 22-year-period from 1988 to 2010. BEAST is designed to reconstruct evolutionary history within the context of geographic distribution over time from sampled DNA sequences16 and has been used extensively in bacterial17,18,19,20, viral21,22 and eukaryotic23 population studies. From this analysis, a single maximum clade credibility (MCC) tree was produced for each lineage (Fig. 2a,b). The mean evolutionary rates, assuming a Bayesian skyline model of population size change and a relaxed molecular clock, were estimated to be 1.9 × 10−7 and 3.9 × 10−7 substitutions per site per year for lineages II and I, respectively. These estimates correspond to an accumulation of approximately 1–2 SNPs per genome per year, which is similar to the substitution rate calculated for the enteric pathogen Vibrio cholerae (8 × 10−7 substitutions per site per year)24 and lies between the rates estimated for Yersinia pestis (2 × 10−8)25 and Staphylococcus aureus (3 × 10−6)26. The topologies of the BEAST and maximum-likelihood trees were congruent, and the recovered nodes were supported with high posterior probabilities and bootstrap values, respectively.
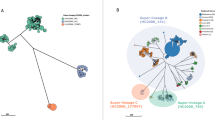
(a,b) MCC trees from BEAST showing phylogeographic reconstruction of lineage I (a) and lineage II (b) with estimated sampling intervals of 43.0 years (1960.6–2003.6) and 32.3 years (1977.1–2009.4), respectively. Estimated ages of nodes where transmissions occurred (black circles) are reported as the median values, with 95% HPD given in parentheses. Asterisk indicates the second introduction of invasive Salmonella Typhimurium in Mali. Posterior probability values for all geographic locations at the ancestral nodes other than the second spread into Uganda (0.56) of >0.9 for lineage I in a and >0.7 for lineage II in b were recovered for all the geographic locations at the ancestral nodes. Branches and nodes are colored according to the location that had the highest posterior probability value. Arrows indicate the estimated points of insertion of independently acquired Tn21 and cat loci within the plasmids in both lineages. (c) Percentage of HIV prevalence in sampled countries from 1960 to the present. HIV prevalence is defined as the percentage of men and women between the ages of 15 and 49 who are HIV positive (UNAIDS Report on the Global AIDS Epidemic 2010; see URLs). Dashed lines show predicted HIV prevalence values before monitoring and reportage for the different countries extrapolated backward in time to 1960. The block outlined by dashed lines indicates the time when HIV prevalence monitoring was temporally associated with the expansion of invasive Salmonella Typhimurium clones across sub-Saharan Africa.
A time-dependent phylogeographic reconstruction of lineage I, which is estimated to have emerged ∼52 years ago (95% highest posterior density (HPD) 1920.4–1979.5; Fig. 2a), indicated that, in our collection, isolates from Malawi diverged earliest from the last common ancestor for this lineage. Although we cannot completely eliminate potential bias due to the number of Malawi isolates analyzed within this lineage, 25-permutation data sets using 10 randomly selected Malawi isolates (with a different set of 10 isolates, equivalent to the sample sizes for other countries, used for each permutation) returned similar results to the complete data set. Thus, we are confident of our estimates of the age and geographic origin of the ancestral node of this lineage (Supplementary Fig. 2a,b). Analyses of the distribution of isolates from each country and the tree topology of lineage I are consistent with at least four independent transmission events or movements across southeastern Africa, with Malawi having served as a potentially important early hub (Fig. 3a and Supplementary Fig. 3a). The earliest identifiable waves or transmissions were from Malawi to Kenya in ∼1982 (95% HPD 1967.6–1990.2) and between Malawi and the DRC in ∼1983 (95% HPD 1974.8–1988.3). This same phylogenetically linked wave was present in Uganda in ∼1989 (95% HPD 1980.0–1994.6), and a further outward wave was identifiable in Mozambique in ∼1990 (95% HPD 1981.0–1994.4) and manifested as a second introduction into Uganda in ∼2001 (95% HPD 1981.0–1994.4). We cannot identify the specific geographic route that these bacterial lineages followed, but the phylogenetic evidence clearly temporally links these outbreaks as a single epidemic. Our results also show evidence of geographic clustering after a transmission event introduced the lineage into a country. This suggests that the epidemic clone was introduced a limited number of times into each country, giving rise to localized epidemics or outbreaks.
(a,b) Phylogeographic diffusion of lineages I (a) and II (b) across sub-Saharan Africa over time based on a discrete geospatial model with associated geographic coordinates. Countries shown here represent discrete locations annotated at the tree nodes taken from the BEAST analyses, and branches that indicate location changes are represented on the map as the transmission lines. The color gradient shows the ages of transmission lines.
Invasive Salmonella Typhimurium isolates of lineage I disappeared from our collection between 2003 and 2005 and were replaced by isolates from lineage II, with isolates from after 2006 found exclusively in this cluster. Lineage II is estimated to have emerged ∼35 years ago (95% HPD 1957.1–1986.8), making it genetically younger than lineage I (Fig. 2b). The spread of lineage II also seems to have occurred in several waves (Fig. 3b and Supplementary Fig. 3b). Our deepest-rooted isolates are from the DRC, with evidence for transmission outward to Uganda in ∼1985 (95% HPD 1972.6–1990.6). This wave was detected in Kenya and Malawi between 1994 and 1996. Malawi likely represents a more recent hub for further dispersal of invasive Salmonella Typhimurium lineage II isolates between 1995 and 1998 to several countries, including neighboring Mozambique, and reaching further westward, across the sub-Saharan region, to Mali and Nigeria. A more recent wave of this lineage seems to have spread from Kenya, arriving back in Malawi in ∼2002. We also detected evidence of localized epidemics associated with the lineage II clones, as highlighted by clustering based on geography. Indeed, local epidemiology and molecular typing in Malawi and Kenya7,8 of invasive Salmonella Typhimurium isolates from 1997 to 2006 describe a local clonal replacement event of lineage I by lineage II that was associated with the emergence of chloramphenicol resistance in an 18-month period from 2001 to 2003.
Evolution of MDR and potential role of cat gene in clonal replacement
Previously, we characterized two distinct composite Tn21-like transposition elements encoding MDR determinants located on the so-called virulence-associated plasmid pSLT in two representative invasive Salmonella Typhimurium isolates, A130 (lineage I) and D23580 (lineage II)7. These Tn21 elements are inserted at different sites in the pSLT virulence plasmid in each isolate. Notably, in our phylogenetic analysis, we found these insertion sites to be identical within each lineage but different between lineages, suggesting that Tn21 element acquisition was an independent and early event in each lineage (Fig. 4 and Supplementary Fig. 4). Only one isolate from lineage I (A24924) and one isolate from lineage II (254DRC) did not have a Tn21-like element (Fig. 4). Comparative analyses of these two isolates, which are significantly the most deeply rooted isolates in each lineage, showed that, although the relevant variant of the Tn21 element is absent in both isolates (Fig. 2a,b), they share the pSLT plasmid backbone with other isolates of the same lineage. This finding suggests that each shares a common ancestor with the other isolates within the same lineage, with this ancestor having existed before the acquisition of the composite Tn21-like elements (Supplementary Note). With the exception of a deletion in istA—a transposase of insertion sequence IS1326—in A16083, the lineage I–specific Tn21 locus is relatively highly conserved in most isolates of lineage I (Fig. 4b). In contrast, the Tn21-like locus encoded by lineage II isolates seems to be somewhat unstable, as isolates in different parts of the tree (14DRC, 5582, J17 and A32751) have lost subsets of genes (Fig. 4a and Supplementary Fig. 5).
(a) Tn21 elements in lineage I and lineage II mapped to Tn21 element from strain D23580 (top row). (b) Lineage I elements mapped to assembled Tn21 sequence in strain A130 (top row). Sequence reads mapping to the complete sequence length are represented as a heatmap, with dark green color indicating >90% (high) coverage, light green indicating coverage of >30 and <90% and white indicating <30% (low) coverage.
One notable feature of the data set is the absence of a chloramphenicol resistance (cat) gene in all isolates in lineage I. In contrast, the gene was present in >97% of lineage II isolates, with only two isolates lacking it (Fig. 4a). These two isolates are 254DRC, which does not have a Tn21 element, and 5582, a 2005 Kenya isolate where the cat gene was lost due to a simple deletion event (Fig. 4a). These observations strongly suggest the independent acquisition of the cat gene, carried on a lineage II–specific Tn21 element, early on in the genealogy, most likely around the time of expansion from the DRC, as shown in Figure 2b (median node date 1984, 95% HPD 1972.6–1990.6; state posterior probability = 0.78). The analysis of MDR acquisition is consistent with the antibiotic resistance profiles obtained for the isolates. In some of our sampling sites, such as Malawi, the acquisition of resistance to chloramphenicol was observed in invasive Salmonella Typhimurium isolates from around 2001–2004, consistent with the arrival of lineage II clones7. At this time, chloramphenicol was the drug of choice for treatment of suspected severe bacterial infections and cases of iNTS infection confirmed by blood culture. The acquisition of chloramphenicol resistance may have afforded lineage II clones a greater opportunity to survive treatment and transmit, which could have in turn contributed to the clonal replacement of lineage I strains, as observed between 2003 and 2005, and the expansion of lineage II clones thereafter.
Transmission is temporally associated with HIV and the HIV pandemic
Time-dependent phylogeographic analysis identified the clonal expansion of two distinct invasive Salmonella Typhimurium lineages within the last 40–50 years that was accompanied by spread across multiple countries of sub-Saharan Africa. Notably, this emergence temporally coincides with the HIV pandemic in sub-Saharan Africa. Molecular clock analysis of HIV-1 genome sequences suggested that the pandemic began at the start of the twentieth century27,28,29, with prevalence peaking in the 1990s in many countries, including those represented within our strain collection (from 2% in Mali to over 15% in Malawi) (Fig. 2c and Supplementary Fig. 6). Association with the HIV status of the affected individuals is also reflected in terms of the samples analyzed in this study. For example, where a test was conducted for HIV, all adult samples were positive. One of the first reported cases of HIV infection in Africa was from an adult in the DRC30, and, notably, the earliest geographic localization of epidemic clones from lineage II was within this country. Thus, the Congo basin represents a potential origin of invasive Salmonella Typhimurium lineage II (ref. 31). It therefore seems possible that the epidemic of invasive Salmonella Typhimurium and transmission across the sub-Saharan region were potentiated by an increase in the critical population of susceptible and immunocompromised individuals, in particular, more mobile adults.
Discussion
The recent reporting of a very high incidence of invasive Salmonella Typhimurium in various parts of the sub-Saharan African region makes it increasingly important to understand the evolutionary origins and spatiotemporal spread of these isolates. Recently, whole-genome sequencing methods have been used to trace intercontinental transmission of different recently emerged and closely related bacterial pathogens18,24,26,32, and we have therefore applied this high-resolution analysis to determine the phylogenetic structure of invasive Salmonella Typhimurium. Here, we find that the vast majority of Salmonella Typhimurium isolates associated with invasive disease from sub-Saharan Africa comprised just two highly conserved lineages of MLST group ST313 that are more closely related to each other than any other known Salmonella Typhimurium lineage. This is in contrast to the considerable phylogenetic variation of the Salmonella Typhimurium isolates associated with gastroenteritis or invasive disease from outside sub-Saharan Africa. Thus, invasive Salmonella Typhimurium–mediated disease in this region is in part a previously unrecognized epidemic caused by the spread of the clones from these two lineages.
We show how invasive Salmonella Typhimurium transmission into a particular country or geographic area occurs as a discrete, temporally defined introduction that is followed by subsequent spread within that particular location (Fig. 2), although some local regions have experienced multiple introduction events. For example, it is evident that two independent introduction events occurred in Mali between 1995 and 2000 (Fig. 2b). Considerable clonal expansion has occurred independently in each of these two lineages, beginning around 1960. Independent acquisition of a Tn21 element encoding MDR genes by both lineages may have facilitated their successful transmission across the subcontinent within the susceptible host population. A later acquisition of a cat gene on the composite element within lineage II has contributed to a clonal replacement event, which occurred between 2003 and 2005 and resulted in greater spatial dispersion of clones from this lineage over sub-Saharan Africa. An association between acquisition of chloramphenicol resistance and increased transmission has been observed in early epidemiological studies on chloramphenicol-resistant Salmonella Typhi in Mexico33 and is also confirmed by observations reported in Kenya7 and Malawi8.
HIV increases susceptibility to iNTS infections34, and this form of bacteremia is an AIDS-defining opportunistic infection in adults35,36. Further, animal models of co-infection with iNTS strains and simian immunodeficiency virus (SIV)37 or malaria38 indicate that host immune status has a critical role in determining the outcome of Salmonella infections. Indeed, sporadic human invasive disease is a feature of the non-ST313 lineages of Salmonella Typhimurium. Thus, although ST313 is the dominant form of invasive Salmonella disease in sub-Saharan Africa3,39, it is not unexpected that other S. enterica or indeed Salmonella Typhimurium lineages can also cause sporadic disease. Notably, supporting epidemiological evidence indicates that the ST313 Salmonella Typhimurium lineages may not have reached some parts of Africa, including the Gambia40,41 and Ethiopia42,43, where iNTS has been reported.
It is particularly noteworthy that we see a temporal association of clonal expansion of invasive Salmonella Typhimurium with the peaks in HIV prevalence, particularly in adults in the countries included in our study. The rapid expansion and spread of these clones may have been facilitated by the dramatic expansion of a mobile susceptible host population. Previous analysis has shown that HIV-I arrived in east and central Africa around the 1950s and expanded eastward in the 1970s and early 1980s (ref. 44). We find temporal parallels in this estimated HIV-I expansion timeframe and our estimate of the earliest detectable transmissions in lineage I around the early 1980s (95% HPD 1967.6–1990.2). The continued expansion of the HIV-susceptible population until the peaks of prevalence in the 1990s (Fig. 2c), together with the acquisition of additional chloramphenicol resistance, is likely contributory to the greater dispersal of lineage II clones. The association of iNTS disease with malaria, anemia and malnourishment in children is well documented4,5,45,46,47, and we have isolates within our collection from children with these underlying conditions (Supplementary Table 1). Malnourished and malarial children thus present an additional ecological niche that coexists with as well as precedes the HIV-positive population. Notably, we found no evidence of phylogenetic segregation between such isolates and those from HIV-positive children or adults within the two epidemic lineages. This is consistent with immunosuppression being a key predisposing factor in iNTS disease. However, the emergence of a large cohort of HIV-infected adults may also have facilitated the spread of the invasive Salmonella Typhimurium lineages, as adults are inevitably more mobile than children. This is especially pertinent because failure of immunological control of iNTS infections in HIV-positive African adults has been well documented34,48.
The resulting large pool of immunosuppressed individuals may also facilitate an unusual human-to-human transmission (anthroponotic) component in invasive Salmonella Typhimurium disease, in contrast to most disease caused by NTS outside of Africa, where transmission is predominantly zoonotic49. There is a dearth of information on the specifics of NTS transmission in sub-Saharan Africa, although independent, country-based studies have shown evidence of non-zoonotic transmission patterns39,49,50. It is perhaps noteworthy that we detected a similar pattern of genomic degradation in the form of gene loss and pseudogene formation to that seen in the human-adapted Salmonella serovars Typhi12 and Paratyphi51 in the two fully sequenced African invasive Salmonella Typhimurium isolates, D23580 and A130, which are representative of lineages I and II, respectively7. Taken together, these results suggest that the invasive clones may have adapted to facilitate direct person-to-person transmission within the human population. Further comparative studies on the virulence and transmission potential of different Salmonella Typhimurium lineages will be instrumental in closing this critical knowledge gap and are the focus of ongoing investigations.
These results provide the first whole genome–based transmission study of this kind on iNTS isolates from sub-Saharan Africa, and they highlight the power of these approaches to monitor the emergence and spread over time of clonal bacterial populations associated with epidemics locally or globally. The transmission pathways hypothesized here suggest potential routes to the implementation of appropriate clinical intervention strategies.
URLs.
European Nucleotide Archive (ENA), http://www.ebi.ac.uk/ena/; MLST database, http://mlst.ucc.ie/mlst/mlst/dbs/Senterica/; AIDSInfoOnline.mdb, http://www.aidsinfoonline.org/; UNAIDS, http://www.unaids.org/en/; UNAIDS Report on the Global AIDS Epidemic 2010, http://www.unaids.org/globalreport/global_report.htm; Google Earth, http://www.google.co.uk/intl/en_uk/earth/index.html.
Methods
Isolate selection and genomic DNA preparation.
We cultured 129 isolates associated with invasive disease from Malawi, Mali, Kenya and Nigeria from the venous blood, cerebrospinal fluid or stool of febrile adults and children between 1988 and 2010. Gastrointestinal isolates were obtained from collections at the Salmonella Genetic Stock center (SGSC)52, the Health Protection Agency or as indicated in Supplementary Table 1 (refs. 13,53,54,55,56,57). Invasive Salmonella Typhimurium isolates were identified by standard serotyping methods, using O- and H-antigen agglutination, based on the Kauffmann-White Scheme1. DNA samples were provided for invasive Salmonella Typhimurium isolates from the DRC, Mozambique and Uganda. Isolates were grown on LB medium, and single colonies were incubated in LB broth overnight at 37 °C. Bacterial cells were pelleted by centrifugation (3,700 g (4,300 rpm) for 5 min), and DNA was extracted using either the Wizard Genomic DNA kit (Promega) according to the manufacturer's instructions or a phenol/chloroform extraction protocol18. DNA quality and quantity were evaluated by gel electrophoresis and the Qubit quantitation platform (Invitrogen). We submitted 20–50 ng/μl DNA from each isolate for Illumina sequencing.
Genomic library preparation and sequencing.
Multiplex libraries with a 200-bp insert size were prepared using 12 unique index tags and were sequenced to generate 54- or 76-bp paired-end reads. Cluster formation, primer hybridization and sequencing reactions were based on reversible terminator chemistry using the Illumina Genome Analyzer II system according to standard protocols26,58. Sequence data were submitted to the European Nucleotide Archive (the full list of accession codes is given in Supplementary Table 1).
Read alignment and SNP detection.
Paired-end Illumina sequence data from each isolate were mapped to the reference genome of the Salmonella Typhimurium strain SL1344 (ref. 57) using SSAHA2 (ref. 59). Sequence reads mapped to an average of 97.7% of the reference genome, with a mean depth of 56.5-fold in mapped regions across all isolates (Supplementary Table 1). SNPs were identified using SAMtools mpileup and were filtered for a minimum mapping quality of 30 and a quality ratio cutoff of 0.75 (refs. 18,24,26,59,60). SNPs called in phage sequences and repetitive regions of the Salmonella Typhimurium reference genome were excluded. Repetitive regions were defined as exact repetitive sequences of ≥20 bp in length, identified using repeat-finding programs NUCmer61, REPeuter62 and repeat-match12,17. Recombinant segments of the genome were removed from the whole-genome alignment as described previously18. After the removal of recombinant segments, mobile elements and repetitive sequences, a concatenated alignment composed of 10,623 SNP sites from each sequenced isolate was produced. Small insertions and deletions (indels) were also identified from the SSAHA result output but were not used for subsequent phylogenetic analyses.
Phylogenetic analyses.
A maximum-likelihood phylogenetic tree (Fig. 1) was constructed from SNP alignment with RAxML v7.0.4 (ref. 14) using a general time-reversible (GTR) substitution model with γ correction for among-site rate variation. Support for nodes on the trees was assessed using 100 bootstrap replicates. For the identified lineages I and II, 487 and 422 chromosomal SNP loci were identified, respectively. These within-cluster SNP alignments were then used to recalculate individual maximum-likelihood trees for each cluster, using the same parameters. These trees were used as input for subsequent analyses. These methods were also applied to obtain a maximum-likelihood phylogenetic reconstruction of plasmids from our isolate collection using 1,251 concatenated SNP sites with the virulence plasmid pSLT-SL1344 from SL1344 as the reference.
MLST analyses.
Allele coordinates were obtained for the seven housekeeping genes used for the S. enterica MLST typing scheme (aroC, dnaN, hemD, hisD, purE, sucA and thrA) by manually marking the coordinates in the whole-genome alignments of our isolates. The marked regions were extracted, and a multisequence alignment was produced for each gene for all the isolates. The resulting alignments were used to determine the sequence type of each isolate using the S. enterica MLST database.
Bayesian phylogeny, estimating dates of divergence and phylogeographic analyses of lineages.
Estimation of rates of evolution, divergence times and phylogeography for our isolate collection as well as for each of the identified lineages was performed using the Bayesian MCMC framework, BEAST15, on SNP alignments. Various combinations of population size change model and molecular clock model were compared to find the model that best fit the data. In all cases, Bayes factors showed strong support (Bayes factor << 200) for the use of a skyline63 model of population size change and a relaxed uncorrelated lognormal clock64, which allows the evolutionary rates to change among the branches of the tree24, and a GTR substitution model with γ correction for among-site rate variation.
Using the same parameters, the geographic locations of ancestral nodes were estimated using the discrete geospatial model implemented in BEAST (Supplementary Table 1)16. In all cases, 3 independent chains were run for 250 million steps each and were sampled every 10,000 steps. The 3 chains were combined with LogCombiner15 with the initial 25 million steps removed from each as a burn-in. MCC trees were created and annotated using TreeAnnotator and were viewed in FigTree15. We report estimates as median values within 95% HPD and report posterior probability values as support for identified ancestral node age and geographic location. For the latter, we report values greater than 0.7. Spatial reconstruction of MCC trees was carried out using SPREAD software65 and visualized with Google Earth (Supplementary Fig. 3).
HIV prevalence data extrapolation.
HIV prevalence data for the sampled countries were modeled with a generalized logistic (or Richards')66 curve using the grofit R package67. Curves were fit to all data points from the beginning of monitoring until stabilization or decline of the HIV-positive population. We then used these fitted models to extrapolate possible past population sizes.
Validation tests for the origin of lineage I.
We used 25 permutation data sets made up of 10 randomly selected Malawi isolates together with the 7 DRC, 8 Kenya, 8 Mozambique and 7 Uganda isolates to reconstruct Bayesian MCC phylogenetic trees. Each of the 25 data sets included a different set of 10 randomly selected Malawi isolates. The same parameters described above were applied in making the trees. Malawi was the ancestral state of all resulting 25 MCC trees with posterior probability values ranging from 0.58–0.92. The resulting phylogenetic trees and their root location state probability distributions are shown in Supplementary Figure 2b.
Plasmid sequence analyses.
Paired-end sequence reads of each isolate were mapped to multi-fasta sequence features, including the Tn21 locus of pSLT-BT, the reference plasmid from invasive strain D23580, using Burrows-Wheeler Aligner (BWA) software68 with minimum base call quality of 50, minimum mapping quality of 30 and minimum read depth of 4. Isolates from each of the three clusters were analyzed separately by cluster. Isolates with <30% of reads mapping to the length of the feature were interpreted as not having the feature, and those with >70% of reads mapping to the feature were interpreted as having the region of interest. A heatmap of the analysis based on the selected cutoff values was generated and aligned to the BEAST MCC tree of each cluster.
De novo sequence assembly and plasmid genome comparisons.
Paired-end Illumina sequence data were assembled de novo using Velvet69, and parameters were optimized to give the highest N50 value. The multi-contig draft genomes generated for each isolate were ordered using either pSLT or pSLT-BT to confirm plasmid structure using Abacas70. Draft plasmid genomes were used to query pSLT and/or pSLT-BT sequences using BLASTN71, and comparison files were generated and viewed using the Artemis Comparison Tool (ACT)72.
Accession codes.
Referenced accession codes for data deposited in the NCBI Nucleotide database include FQ312003, FN424405, HE654726, FN432031 and AE006471. The full set of primary accession codes for the Illumina sequence reads of 177 invasive and gastrointestinal Salmonella Typhimurium is given in Supplementary Table 1.
References
Popoff, M.Y., Bockemuhl, J. & Gheesling, L.L. Supplement 2002 (no. 46) to the Kauffmann-White scheme. Res. Microbiol. 155, 568–570 (2004).
Langridge, G.C., Nair, S. & Wain, J. Nontyphoidal Salmonella serovars cause different degrees of invasive disease globally. J. Infect. Dis. 199, 602–603 (2009).
Reddy, E.A., Shaw, A.V. & Crump, J.A. Community-acquired bloodstream infections in Africa: a systematic review and meta-analysis. Lancet Infect. Dis. 10, 417–432 (2010).
Graham, S.M. Nontyphoidal salmonellosis in Africa. Curr. Opin. Infect. Dis. 23, 409–414 (2010).
Berkley, J.A. et al. HIV infection, malnutrition, and invasive bacterial infection among children with severe malaria. Clin. Infect. Dis. 49, 336–343 (2009).
Gordon, M.A. et al. Non-typhoidal Salmonella bacteraemia among HIV-infected Malawian adults: high mortality and frequent recrudescence. AIDS 16, 1633–1641 (2002).
Kingsley, R.A. et al. Epidemic multiple drug resistant Salmonella Typhimurium causing invasive disease in sub-Saharan Africa have a distinct genotype. Genome Res. 19, 2279–2287 (2009).
Gordon, M.A. et al. Epidemics of invasive Salmonella enterica serovar Enteritidis and S. enterica Serovar Typhimurium infection associated with multidrug resistance among adults and children in Malawi. Clin. Infect. Dis. 46, 963–969 (2008).
Gordon, M.A. Salmonella infections in immunocompromised adults. J. Infect. 56, 413–422 (2008).
Cheesbrough, J.S., Taxman, B.C., Green, S.D., Mewa, F.I. & Numbi, A. Clinical definition for invasive Salmonella infection in African children. Pediatr. Infect. Dis. J. 16, 277–283 (1997).
Parkhill, J. et al. Complete genome sequence of a multiple drug resistant Salmonella enterica serovar Typhi CT18. Nature 413, 848 (2001).
Holt, K.E. et al. High-throughput sequencing provides insights into genome variation and evolution in Salmonella Typhi. Nat. Genet. 40, 987–993 (2008).
Beltran, P. et al. Reference collection of strains of the Salmonella typhimurium complex from natural populations. J. Gen. Microbiol. 137, 601–606 (1991).
Stamatakis, A. RAxML-VI-HPC: maximum likelihood–based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics 22, 2688–2690 (2006).
Drummond, A.J. & Rambaut, A. BEAST: Bayesian evolutionary analysis by sampling trees. BMC Evol. Biol. 7, 214 (2007).
Lemey, P., Rambaut, A., Drummond, A.J. & Suchard, M.A. Bayesian phylogeography finds its roots. PLOS Comput. Biol. 5, e1000520 (2009).
He, M. et al. Evolutionary dynamics of Clostridium difficile over short and long time scales. Proc. Natl. Acad. Sci. USA 107, 7527–7532 (2010).
Croucher, N.J. et al. Rapid pneumococcal evolution in response to clinical interventions. Science 331, 430–434 (2011).
Holt, K.E. et al. Temporal fluctuation of multidrug resistant Salmonella Typhi haplotypes in the Mekong river delta region of Vietnam. PLoS Negl. Trop. Dis. 5, e929 (2011).
den Bakker, H.C., Bundrant, B.N., Fortes, E.D., Orsi, R.H. & Wiedmann, M. A population genetics–based and phylogenetic approach to understanding the evolution of virulence in the genus Listeria. Appl. Environ. Microbiol. 76, 6085–6100 (2010).
Smith, G.J. et al. Origins and evolutionary genomics of the 2009 swine-origin H1N1 influenza A epidemic. Nature 459, 1122–1125 (2009).
Smith, G.J. et al. Dating the emergence of pandemic influenza viruses. Proc. Natl. Acad. Sci. USA 106, 11709–11712 (2009).
Endicott, P., Ho, S.Y. & Stringer, C. Using genetic evidence to evaluate four palaeoanthropological hypotheses for the timing of Neanderthal and modern human origins. J. Hum. Evol. 59, 87–95 (2010).
Mutreja, A. et al. Evidence for several waves of global transmission in the seventh cholera pandemic. Nature 477, 462–465 (2011).
Morelli, G. et al. Yersinia pestis genome sequencing identifies patterns of global phylogenetic diversity. Nat. Genet. 42, 1140–1143 (2010).
Harris, S.R. et al. Evolution of MRSA during hospital transmission and intercontinental spread. Science 327, 469 (2010).
Korber, B. et al. Timing the ancestor of the HIV-1 pandemic strains. Science 288, 1789–1796 (2000).
Lemey, P. et al. The molecular population genetics of HIV-1 group O. Genetics 167, 1059–1068 (2004).
Worobey, M. et al. Direct evidence of extensive diversity of HIV-1 in Kinshasa by 1960. Nature 455, 661–664 (2008).
Nahmias, A.J. et al. Evidence for human infection with an HTLV III/LAV-like virus in Central Africa, 1959. Lancet 1, 1279–1280 (1986).
Sharp, E.R. et al. Immunodominance of HIV-1 specific CD8+ T-cell responses is related to disease progression rate in vertically infected adolescents. PLoS ONE 6, e21135 (2011).
Harris, S.R. et al. Whole-genome analysis of diverse Chlamydia trachomatis strains identifies phylogenetic relationships masked by current clinical typing. Nat. Genet. 44, 413–419 (2012).
Gangarosa, E.J. et al. An epidemic-associated episome? J. Infect. Dis. 126, 215–218 (1972).
MacLennan, C.A. et al. Dysregulated humoral immunity to nontyphoidal Salmonella in HIV-infected African adults. Science 328, 508–512 (2010).
Smith, P.D. et al. Salmonella typhimurium enteritis and bacteremia in the acquired immunodeficiency syndrome. Ann. Intern. Med. 102, 207–209 (1985).
Levine, W.C., Buehler, J.W., Bean, N.H. & Tauxe, R.V. Epidemiology of nontyphoidal Salmonella bacteremia during the human immunodeficiency virus epidemic. J. Infect. Dis. 164, 81–87 (1991).
Raffatellu, M. et al. Simian immunodeficiency virus–induced mucosal interleukin-17 deficiency promotes Salmonella dissemination from the gut. Nat. Med. 14, 421–428 (2008).
Roux, C.M. et al. Both hemolytic anemia and malaria parasite–specific factors increase susceptibility to Nontyphoidal Salmonella enterica serovar Typhimurium infection in mice. Infect. Immun. 78, 1520–1527 (2010).
Keddy, K.H. et al. Genotypic and demographic characterization of invasive isolates of Salmonella Typhimurium in HIV co-infected patients in South Africa. J. Infect. Dev. Ctries. 3, 585 (2009).
Ikumapayi, U.N. et al. Molecular epidemiology of community-acquired invasive non-typhoidal Salmonella among children aged 2–29 months in rural Gambia and discovery of a new serovar, Salmonella enterica Dingiri. J. Med. Microbiol. 56, 1479 (2007).
Dione, M.M. et al. Clonal differences between Non-Typhoidal Salmonella (NTS) recovered from children and animals living in close contact in the Gambia. PLoS Negl. Trop. Dis. 5, e1148 (2011).
Beyene, G. et al. Multidrug resistant Salmonella Concord is a major cause of salmonellosis in children in Ethiopia. J. Infect. Dev. Ctries. 5, 23–33 (2011).
Sibhat, B. et al. Salmonella serovars and antimicrobial resistance profiles in beef cattle, slaughterhouse personnel and slaughterhouse environment in Ethiopia. Zoonoses Public Health 58, 102–109 (2011).
Gray, R.R. et al. Spatial phylodynamics of HIV-1 epidemic emergence in east Africa. AIDS 23, F9–F17 (2009).
Brent, A.J. et al. Salmonella bacteremia in Kenyan children. Pediatr. Infect. Dis. J. 25, 230–236 (2006).
Mandomando, I. et al. Invasive non-typhoidal Salmonella in Mozambican children. Trop. Med. Int. Health 14, 1467 (2009).
Rosanova, M.T., Paganini, H., Bologna, R., Lopardo, H. & Ensinck, G. Risk factors for mortality caused by nontyphoidal Salmonella sp. in children. Int. J. Infect. Dis. 6, 187–190 (2002).
Gordon, M.A. et al. Invasive non-typhoid salmonellae establish systemic intracellular infection in HIV-infected adults: an emerging disease pathogenesis. Clin. Infect. Dis. 50, 953–962 (2010).
Kariuki, S. et al. Invasive multidrug-resistant non-typhoidal Salmonella infections in Africa: zoonotic or anthroponotic transmission? J. Med. Microbiol. 55, 585–591 (2006).
Fashae, K., Ogunsola, F., Aarestrup, F.M. & Hendriksen, R.S. Antimicrobial susceptibility and serovars of Salmonella from chickens and humans in Ibadan, Nigeria. J. Infect. Dev. Ctries. 4, 484–494 (2010).
Holt, K.E. et al. Pseudogene accumulation in the evolutionary histories of Salmonella enterica serovars Paratyphi A and Typhi. BMC Genomics 10, 36 (2009).
Zinder, N.D. & Lederberg, J. Genetic exchange in Salmonella. J. Bacteriol. 64, 679–699 (1952).
Helm, R.A. et al. Pigeon-associated strains of Salmonella enterica serovar Typhimurium phage type DT2 have genomic rearrangements at rRNA operons. Infect. Immun. 72, 7338 (2004).
Beltran, P. et al. Toward a population genetic analysis of Salmonella: genetic diversity and relationships among strains of serotypes S. choleraesuis, S. derby, S. dublin, S. enteritidis, S. heidelberg, S. infantis, S. newport, and S. typhimurium. Proc. Natl. Acad. Sci. USA 85, 7753–7757 (1988).
Cooke, F.J. et al. Characterization of the genomes of a diverse collection of Salmonella enterica serovar Typhimurium definitive phage type 104. J. Bacteriol. 190, 8155 (2008).
Andrews-Polymenis, H.L. et al. Host restriction of Salmonella enterica serotype Typhimurium pigeon isolates does not correlate with loss of discrete genes. J. Bacteriol. 186, 2619 (2004).
Hoiseth, S.K. & Stocker, B.A. Aromatic-dependent Salmonella typhimurium are non-virulent and effective as live vaccines. Nature 291, 238–239 (1981).
Bentley, D.R. et al. Accurate whole human genome sequencing using reversible terminator chemistry. Nature 456, 53–59 (2008).
Ning, Z., Cox, A.J. & Mullikin, J.C. SSAHA: a fast search method for large DNA databases. Genome Res. 11, 1725–1729 (2001).
Li, H. et al. The Sequence Alignment/Map format and SAMtools. Bioinformatics 25, 2078–2079 (2009).
Kurtz, S. et al. Versatile and open software for comparing large genomes. Genome Biol. 5, R12 (2004).
Kurtz, S. et al. REPuter: the manifold applications of repeat analysis on a genomic scale. Nucleic Acids Res. 29, 4633–4642 (2001).
Drummond, A.J., Rambaut, A., Shapiro, B. & Pybus, O.G. Bayesian coalescent inference of past population dynamics from molecular sequences. Mol. Biol. Evol. 22, 1185–1192 (2005).
Drummond, A.J., Ho, S.Y., Phillips, M.J. & Rambaut, A. Relaxed phylogenetics and dating with confidence. PLoS Biol. 4, e88 (2006).
Bielejec, F., Rambaut, A., Suchard, M.A. & Lemey, P. SPREAD: spatial phylogenetic reconstruction of evolutionary dynamics. Bioinformatics 27, 2910–2912 (2011).
Richards, F.J. A flexible growth function for empirical use. J. Exp. Bot. 10, 290–301 (1959).
Kahm, M., Hasenbrink, G., Lichtenberg-Fraté, H., Ludwig, J. & Kschischo, M. grofit: fitting biological growth curves with R. J. Stat. Softw. 33 (2010).
Li, H. & Durbin, R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25, 1754–1760 (2009).
Zerbino, D.R. & Birney, E. Velvet: algorithms for de novo short read assembly using de Bruijn graphs. Genome Res. 18, 821–829 (2008).
Assefa, S., Keane, T.M., Otto, T.D., Newbold, C. & Berriman, M. ABACAS: algorithm-based automatic contiguation of assembled sequences. Bioinformatics 25, 1968–1969 (2009).
Altschul, S.F., Gish, W., Miller, W., Myers, E.W. & Lipman, D.J. Basic local alignment search tool. J. Mol. Biol. 215, 403–410 (1990).
Carver, T. et al. Artemis and ACT: viewing, annotating and comparing sequences stored in a relational database. Bioinformatics 24, 2672–2676 (2008).
Acknowledgements
We thank J. Cheesborough for providing the DRC isolates, M. Okong, N. French and the Medical Research Council, Uganda, for providing the Uganda isolates, S. Nair for providing the Health Protection Agency (HPA) isolates, L. Barquist for modeling the pre-1990 HIV prevalence data and the Sequencing team at the Wellcome Trust Sanger Institute. This work was funded by a Wellcome Trust grant (098051). C.A.M. was supported by a Tropical Research Fellowship from the Wellcome Trust and a Clinical Research Fellowship from GlaxoSmithKline.
Author information
Authors and Affiliations
Contributions
C.K.O. and R.A.K. contributed to collecting data and manuscript writing. C.K.O. analyzed sequence data and performed phylogenetic, BEAST and comparative genomics analyses. T.R.C. and S.R.H. wrote the coding scripts for phylogenetic and Bayesian statistical analyses and contributed to manuscript writing. C.M.P., M.N.A.-M., S.K., C.L.M., M.A.G., E.d.P., R.S.H., S.O., P.L.A., I.M., C.A.M., J.W., M.D.T., M.M.L. and S.M.T. contributed to studies from which isolates were drawn and to manuscript writing. G.D., J.P. and R.A.K. designed the study, and G.D. supervised the work.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–6 and Supplementary Note (PDF 623 kb)
Supplementary Table 1
Isolates used in study with mapping statistics and metadata (to be included as a separate file in Excel format) (XLS 83 kb)
Rights and permissions
About this article
Cite this article
Okoro, C., Kingsley, R., Connor, T. et al. Intracontinental spread of human invasive Salmonella Typhimurium pathovariants in sub-Saharan Africa. Nat Genet 44, 1215–1221 (2012). https://doi.org/10.1038/ng.2423
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ng.2423