Abstract
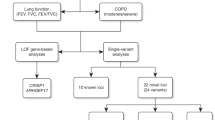
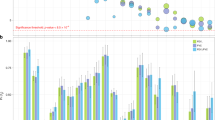
Pulmonary function measures reflect respiratory health and are used in the diagnosis of chronic obstructive pulmonary disease. We tested genome-wide association with forced expiratory volume in 1 second and the ratio of forced expiratory volume in 1 second to forced vital capacity in 48,201 individuals of European ancestry with follow up of the top associations in up to an additional 46,411 individuals. We identified new regions showing association (combined P < 5 × 10−8) with pulmonary function in or near MFAP2, TGFB2, HDAC4, RARB, MECOM (also known as EVI1), SPATA9, ARMC2, NCR3, ZKSCAN3, CDC123, C10orf11, LRP1, CCDC38, MMP15, CFDP1 and KCNE2. Identification of these 16 new loci may provide insight into the molecular mechanisms regulating pulmonary function and into molecular targets for future therapy to alleviate reduced lung function.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout


Similar content being viewed by others
References
Wilk, J.B. et al. Evidence for major genes influencing pulmonary function in the NHLBI family heart study. Genet. Epidemiol. 19, 81–94 (2000).
Hole, D.J. et al. Impaired lung function and mortality risk in men and women: findings from the Renfrew and Paisley prospective population study. Br. Med. J. 313, 711–715, discussion 715–716 (1996).
Strachan, D.P. Ventilatory function, height, and mortality among lifelong non-smokers. J. Epidemiol. Community Health 46, 66–70 (1992).
Young, R.P., Hopkins, R. & Eaton, T.E. Forced expiratory volume in one second: not just a lung function test but a marker of premature death from all causes. Eur. Respir. J. 30, 616–622 (2007).
Lopez, A.D. et al. Chronic obstructive pulmonary disease: current burden and future projections. Eur. Respir. J. 27, 397–412 (2006).
Mathers, C.D. & Loncar, D. Projections of global mortality and burden of disease from 2002 to 2030. PLoS Med. 3, e442 (2006).
Rabe, K.F. et al. Global strategy for the diagnosis, management, and prevention of chronic obstructive pulmonary disease: GOLD executive summary. Am. J. Respir. Crit. Care Med. 176, 532–555 (2007).
Hancock, D.B. et al. Meta-analyses of genome-wide association studies identify multiple loci associated with pulmonary function. Nat. Genet. 42, 45–52 (2010).
Repapi, E. et al. Genome-wide association study identifies five loci associated with lung function. Nat. Genet. 42, 36–44 (2010).
Pillai, S.G. et al. A genome-wide association study in chronic obstructive pulmonary disease (COPD): identification of two major susceptibility loci. PLoS Genet. 5, e1000421 (2009).
Wilk, J.B. et al. A genome-wide association study of pulmonary function measures in the Framingham Heart Study. PLoS Genet. 5, e1000429 (2009).
Yang, J. et al. Genomic inflation factors under polygenic inheritance. Eur. J. Hum. Genet. 19, 807–812 (2011).
Elks, C.E. et al. Thirty new loci for age at menarche identified by a meta-analysis of genome-wide association studies. Nat. Genet. 42, 1077–1085 (2010).
Lango Allen, H. et al. Hundreds of variants clustered in genomic loci and biological pathways affect human height. Nature 467, 832–838 (2010).
Lindgren, C.M. et al. Genome-wide association scan meta-analysis identifies three loci influencing adiposity and fat distribution. PLoS Genet. 5, e1000508 (2009).
Dixon, A.L. et al. A genome-wide association study of global gene expression. Nat. Genet. 39, 1202–1207 (2007).
Gibson, M.A., Hughes, J.L., Fanning, J.C. & Cleary, E.G. The major antigen of elastin-associated microfibrils is a 31-kDa glycoprotein. J. Biol. Chem. 261, 11429–11436 (1986).
Yang, J. et al. Rootletin, a novel coiled-coil protein, is a structural component of the ciliary rootlet. J. Cell Biol. 159, 431–440 (2002).
Massaro, G.D. et al. Retinoic acid receptor-β: an endogenous inhibitor of the perinatal formation of pulmonary alveoli. Physiol. Genomics 4, 51–57 (2000).
Bieganowski, P., Shilinski, K., Tsichlis, P.N. & Brenner, C. Cdc123 and checkpoint forkhead associated with RING proteins control the cell cycle by controlling eIF2γ abundance. J. Biol. Chem. 279, 44656–44666 (2004).
Ito, K. et al. Decreased histone deacetylase activity in chronic obstructive pulmonary disease. N. Engl. J. Med. 352, 1967–1976 (2005).
Wu, H. et al. Parental smoking modifies the relation between genetic variation in tumor necrosis factor-α (TNF) and childhood asthma. Environ. Health Perspect. 115, 616–622 (2007).
Ruse, C.E. et al. Tumour necrosis factor gene complex polymorphisms in chronic obstructive pulmonary disease. Respir. Med. 101, 340–344 (2007).
Chu, H.W. et al. Transforming growth factor-β2 induces bronchial epithelial mucin expression in asthma. Am. J. Pathol. 165, 1097–1106 (2004).
Liu, J.Z. et al. Meta-analysis and imputation refines the association of 15q25 with smoking quantity. Nat. Genet. 42, 436–440 (2010).
Landi, M.T. et al. A genome-wide association study of lung cancer identifies a region of chromosome 5p15 associated with risk for adenocarcinoma. Am. J. Hum. Genet. 85, 679–691 (2009).
Kathiresan, S. et al. Genome-wide association of early-onset myocardial infarction with single nucleotide polymorphisms and copy number variants. Nat. Genet. 41, 334–341 (2009).
Clancy, R.M. et al. Identification of candidate loci at 6p21 and 21q22 in a genome-wide association study of cardiac manifestations of neonatal lupus. Arthritis Rheum. 62, 3415–3424 (2010).
Harley, J.B. et al. Genome-wide association scan in women with systemic lupus erythematosus identifies susceptibility variants in ITGAM, PXK, KIAA1542 and other loci. Nat. Genet. 40, 204–210 (2008).
Zeggini, E. et al. Meta-analysis of genome-wide association data and large-scale replication identifies additional susceptibility loci for type 2 diabetes. Nat. Genet. 40, 638–645 (2008).
Barrett, J.C. et al. Genome-wide association study and meta-analysis find that over 40 loci affect risk of type 1 diabetes. Nat. Genet. 41, 703–707 (2009).
Levy, D. et al. Genome-wide association study of blood pressure and hypertension. Nat. Genet. 41, 677–687 (2009).
Newton-Cheh, C. et al. Genome-wide association study identifies eight loci associated with blood pressure. Nat. Genet. 41, 666–676 (2009).
Park, J.H. et al. Estimation of effect size distribution from genome-wide association studies and implications for future discoveries. Nat. Genet. 42, 570–575 (2010).
Yang, J. et al. Genome partitioning of genetic variation for complex traits using common SNPs. Nat. Genet. 43, 519–525 (2011).
Kohansal, R. et al. The natural history of chronic airflow obstruction revisited: an analysis of the framingham offspring cohort. Am. J. Respir. Crit. Care Med. 180, 3–10 (2009).
Hankinson, J.L., Odencrantz, J.R. & Fedan, K.B. Spirometric reference values from a sample of the general U.S. population. Am. J. Respir. Crit. Care Med. 159, 179–187 (1999).
Talmud, P.J. et al. Utility of genetic and non-genetic risk factors in prediction of type 2 diabetes: Whitehall II prospective cohort study. Br. Med. J. 340, b4838 (2010).
Wacholder, S. et al. Performance of common genetic variants in breast-cancer risk models. N. Engl. J. Med. 362, 986–993 (2010).
Thompson, H.G.R., Mih, J.D., Krasieva, T.B., Tromberg, B.J. & George, S.C. Epithelial-derived TGF-β2 modulates basal and wound-healing subepithelial matrix homeostasis. Am. J. Physiol. Lung Cell. Mol. Physiol. 291, L1277–L1285 (2006).
Maran, C., Tassone, E., Masola, V. & Onisto, M. The story of SPATA2 (spermatogenesis-associated protein 2): from Sertoli cells to pancreatic β-cells. Curr. Genomics 10, 361–363 (2009).
Tewari, R., Bailes, E., Bunting, K.A. & Coates, J.C. Armadillo-repeat protein functions: questions for little creatures. Trends Cell Biol. 20, 470–481 (2010).
Pende, D. et al. Identification and molecular characterization of NKp30, a novel triggering receptor involved in natural cytotoxicity mediated by human natural killer cells. J. Exp. Med. 190, 1505–1516 (1999).
Lillis, A.P., Mikhailenko, I. & Strickland, D.K. Beyond endocytosis: LRP function in cell migration, proliferation and vascular permeability. J. Thromb. haemost. 3, 1884–1893 (2005).
Burkhard, P., Stetefeld, J. & Strelkov, S.V. Coiled coils: a highly versatile protein folding motif. Trends Cell Biol. 11, 82–88 (2001).
Cowley, E.A. & Linsdell, P. Characterization of basolateral K+ channels underlying anion secretion in the human airway cell line Calu-3. J. Physiol. (Lond.) 538, 747–757 (2002).
Li, Y. & Abecasis, G.R. Mach 1.0: rapid haplotype construction and missing genotype inference. Am. J. Hum. Genet. S79, 2290 (2006).
Marchini, J., Howie, B., Myers, S., McVean, G. & Donnelly, P. A new multipoint method for genome-wide association studies by imputation of genotypes. Nat. Genet. 39, 906–913 (2007).
Servin, B. & Stephens, M. Imputation-based analysis of association studies: candidate regions and quantitative traits. PLoS Genet. 3, e114 (2007).
Devlin, B. & Roeder, K. Genomic control for association studies. Biometrics 55, 997–1004 (1999).
Cho, M.H. et al. Variants in FAM13A are associated with chronic obstructive pulmonary disease. Nat. Genet. 42, 200–202 (2010).
Pillai, S.G. et al. Loci identified by genome-wide association studies influence different disease-related phenotypes in chronic obstructive pulmonary disease. Am. J. Respir. Crit. Care Med. 182, 1498–1505 (2010).
Segrè, A.V., Groop, L., Mootha, V.K., Daly, M.J. & Altshuler, D. Common inherited variation in mitochondrial genes is not enriched for associations with type 2 diabetes or related glycemic traits. PLoS Genet. 6, e1001058 (2010).
Raychaudhuri, S. et al. Identifying relationships among genomic disease regions: predicting genes at pathogenic SNP associations and rare deletions. PLoS Genet. 5, e1000534 (2009).
Acknowledgements
We thank the many colleagues who contributed to collection and phenotypic characterization of the clinical sampling, genotyping and analysis of the data. We especially thank those who kindly agreed to participate in the studies.
Major funding for this work is from the following sources (alphabetical): Academy of Finland (project grants 104781, 120315, 129269, 1114194, Center of Excellence in Complex Disease Genetics (213506 and 129680) and SALVE); Althingi (Icelandic Parliament); Arthritis Research Campaign; Asthma UK; AstraZeneca; AXA Research Fund; Biotechnology and Biological Sciences Research Council (BBSRC) (BB/F019394/1, G20234); British Heart Foundation (PG/97012, PG/06/154/22043, FS05/125); British Lung Foundation; Canadian Institutes of Health Research (Grant ID MOP-82893); Cancer Research United Kingdom; Chief Scientist Office, Scottish Government Health Directorate (CZD/16/6); Croatian Institute for Public Health; UK Department of Health; Dutch Kidney Foundation; Erasmus Medical Center and Erasmus University, Rotterdam; Estonian Genome Center, University of Tartu, Estonia (SF0180142s08); EU funding (GABRIEL GRANT Number: 018996, ECRHS II Coordination Number: QLK4-CT-1999-01237); European Commission (DG XII, EURO-BLCS, FP-5 QLG1-CT-2000-01643, FP-6 LSHB-CT-2006-018996 (GABRIEL), FP-6 LSHG-CT-2006-018947 (EUROSPAN), FP-6 GenomEUtwin project QLG2-CT-2002-01254, FP7/2007-2013: HEALTH-F2-2008-201865, GEFOS, HEALTH-F2-2008-35627, TREAT-OA, HEALTH-F4-2007-201413 (ENGAGE)); Finnish Foundation for Cardiovascular Research; Flight Attendant Medical Research Institute (FAMRI); German Asthma and COPD Network (COSYCONET: BMBF grant 01GI0883); German Bundesministerium fuer Forschung und Technology (01 AK 803 A-H, 01 IG 07015 G); German Federal Ministry of Education and Research (BMBF) (03ZIK012, 01ZZ9603, 01ZZ0103 and 01ZZ0403): German National Genome Research Network (NGFN-2 and NGFN-plus); German Ministry of Cultural Affairs; GlaxoSmithKline; Gyllenberg Foundations; Healthway, Western Australia; Helmholtz Zentrum München, German Research Center for Environmental Health, Neuherberg, Germany; Healthcare and Bioscience iNet (funded by the East Midlands Development Agency, partially financed by the European Regional Development Fund, delivered by Medilink East Midlands); Higher Education Funding Council for England (HEFCE); Hjartavernd (Icelandic Heart Association); Innsbruck Medical University; Institute for Anthropological Research in Zagreb; International Osteoporosis Foundation; Intramural Research Program of the NIH, National Institute on Aging and National Institute of Environmental Health Sciences; Jalmari and Rauha Ahokas Foundation; Juvenile Diabetes Research Foundation International (JDRF); Lifelong Health and Wellbeing Initiative (G0700704/84698); Medical Research Council UK (G1000861, G0501942, G0902313, G0000934, G0800582, G0500539, G0600705, PrevMetSyn/SALVE, G9901462); Medical Research Fund of the Tampere University Hospital; Ministry of Science, Education and Sport of the Republic of Croatia (108-1080315-0302); Medical Research Council Human Genetics Unit; Medisearch–The Leicester Medical Research Foundation; Munich Center of Health Sciences (MC Health) as part of LMUinnovativ; National Health and Medical Research Council of Australia (Grant ID 403981 and ID 003209); National Human Genome Research Institute (NHGRI) (U01-HG-004729, U01-HG-004402); National Institute for Health Research (NIHR) Comprehensive Biomedical Research Centres (Guy's & St. Thomas' NHS Foundation Trust in partnership with King's College London and Cambridge University Hospitals NHS Foundation Trust in partnership with the University of Cambridge); Netherlands Genomics Initiative (NGI)/Netherlands Organisation for Scientific Research (NWO) (050-060-810); Netherlands Organization for the Health Research and Development (ZonMw); Netherlands Organization of Scientific Research NOW (1750102007006, 175.010.2005.011, 911-03-012); Northern Netherlands Collaboration of Provinces (SNN); Norwegian University of Science and Technology; Novo Nordisk; Ontario Institute of Cancer Research and Canadian Cancer Society Research Institute (CCSRI 020214); Republic of Croatia Ministry of Science, Education and Sports research grants (108-1080315-0302); Research Institute for Diseases in the Elderly (RIDE) (014-93-015: RIDE2); Research Into Ageing (251); Siemens Healthcare, Erlangen, Germany and the Federal State of Mecklenburg–West Pomerania; Social Ministry of the Federal State of Mecklenburg–West Pomerania; Structure Enhancing Fund (FES) of the Dutch government; Swedish Heart and Lung Foundation grant 20050561; Swedish Research Council for Worklife and Social research (FAS), grants 2001-0263, 2003-0139; Swiss National Science Foundation (grants no. 4026-28099,3347CO-108796, 3247BO-104283, 3247BO-104288, 3247BO-104284, 32-65896.01,32-59302.99, 32-52720.97, 32-4253.94); The Asthma, Allergy and Inflammation Research Trust; The Great Wine Estates of the Margaret River region of Western Australia; The Netherlands' Ministry of Economic Affairs, Ministry of Education, Culture and Science and Ministry for Health, Welfare and Sports; The Royal Society; The University of Split and Zagreb Medical Schools; Tromsø University; U01 DK062418; UBS Wealth Foundation Grant BA29s8Q7-DZZ; UK Department of Health Policy Research Programme; University Hospital Oulu, Biocenter, University of Oulu, Finland (75617); University Medical Center Groningen; University of Bristol; University of Leicester HEFCE CIF award; University of Nottingham; US National Institutes of Health (NIH) (1P50 CA70907, RO1 CA121197, U19 CA148127, CA55769, CA127219, R01HL071051, R01HL071205, R01HL071250, R01HL071251, R01HL071252, R01HL071258, R01HL071259, UL1RR025005, contracts HHSN268200625226C, HHSN268200782096C, R01-HL084099); US NIH National Cancer Institute (RO1CA111703); US NIH National Center for Research Resources (grants M01-RR00425 and 5M01 RR00997); US NIH National Eye Institute (NEI); US NIH National Heart, Lung and Blood Institute (contracts N01-HC-85079 through N01-HC-85086, N01-HC-35129, N01 HC-15103, N01 HC-55222, N01-HC-75150, N01-HC-45133, N01-HC-95095, N01-HC-48047, N01-HC-48048, N01-HC-48049, N01-HC-48050, N01-HC-45134, N01-HC-05187, N01-HC-45205, N01-HC-45204, N01 HC-25195, N01-HC-95159 through N01-HC-95169, RR-024156, N02-HL-6-4278, R01 HL-071022, R01 HL-077612, R01 HL-074104, RC1 HL100543, HHSN268201100005C, HHSN268201100006C, HHSN268201100007C, HHSN268201100008C, HHSN268201100009C, HHSN268201100010C, HHSN268201100011C and HHSN268201100012C, grants HL080295, HL087652, HL105756, R01-HL-084099, R01HL087641, R01HL59367, R01HL086694, HL088133, HL075336 5R01HL087679-02 through the STAMPEED program (1RL1MH083268-01), 1K23HL094531-01); US NIH National Institute of Allergy and Infectious Diseases (NIAID); US NIH National Institute of Child Health and Human Development (NICHD); US NIH National Institute of Diabetes and Digestive and Kidney Diseases (NIDDK) (DK063491); US NIH National Institute of Environmental Health Sciences (NIEHS) (ZO1 ES49019, ES015794); US NIH National Institute of Mental Health (NIMH) (5R01MH63706:02); US NIH National Institute of Neurological Disorders and Stroke (NINDD); US NIH National Institute on Aging (NIA) (R01 AG032098, RC1 AG035835, N01AG12100, N01AG62101, N01AG62103, N01AG62106, 1R01AG032098-01A1, AG-023269, AG-15928, AG-20098, AG-027058); Wellcome Trust (077016/Z/05/Z, GR069224, 068545/Z/02, 076113/B/04/Z, 079895).
Author information
Authors and Affiliations
Consortia
Contributions
Author contributions are listed in alphabetical order. See Supplementary Note for definitions of study acronyms.
Project conception, design and management. Stage 1 GWAS, AGES: G.E., M.G., V.G., T.B.H., L.J.L. ARIC: S.J.L., N.F., L.R.L., D.J.C., D.B.H., B.R.J., A.C.M., K.E.N. B58C-T1DGC: D.P.S. B58C -WTCCC: D.P.S. BHS1: A.L.J., A.W.M., L.J.P. CHS: S.A.G., S.R.H., T.L., B.M.P. CROATIA-Korcula: H.C., I.G., S.J., I.R., A.F.W., L.Z. CROATIA-Vis: H.C., C.H., O.P., I.R., A.F.W. ECRHS: D.L.J., E.O., I.P., M.W. EPIC: N.J.W. FHS: J.B.W., G.T.O. FTC: J.K., K.H.P., T. Rantanen. Health ABC: M.C.A., P.A.C., T.B.H., S.B.K., Y.L., B.M. Health 2000: M.H., M.K. KORA F4: J. Heinrich. KORA S3: C.G., H.-E.W. NFBC1966: P.E., A.-L.H., M.-R.J., A.P. ORCADES: H.C., S.H.W., J.F.W., A.F.W. RS: A. Hofman. SHIP: S.G., G.H., B.K., H.V. TwinsUK: T.D.S., G.Z. Stage 2 follow up, ADONIX: J. Brisman., A.-C.O. BHS2: J. Beilby. BRHS: R.W.M., S.G.W., P.H.W. BWHHS: G.D.S., S.E., D.A.L., P.H.W. CARDIA: A.S. CROATIA-Split: M.B., I.K., T.Z. GS: SFHS: C.M.J., S.M.K., A.D.M., D.J.P. HCS: C.C., J.W.H., A.A.S. LBC1936: I.J.D., S.E.H., J.M.S. LifeLines: H.M.B., D.S.P., J.M.V., C.W. MESA-Lung: R.G.B., J.L.H. Nottingham smokers: I.P.H. NSHD: R.H., D.K. SAPALDIA: N.P.-H., T. Rochat. Look-up studies, ALSPAC: R.G., J. Henderson. ILCCO: ILCCO data. Ox-GSK: C.F., J.M.
Phenotype collection and data management. Stage 1 GWAS, AGES: T.A. ARIC: D.J.C., N.F., L.R.L., A.C.M., K.E.N. B58C-T1DGC: A.R.R., D.P.S. B58C -WTCCC: A.R.R., D.P.S. BHS1: A.L.J., A.W.M., L.J.P. CHS: S.A.G., S.R.H., T.L., B.M.P. CROATIA-Korcula: I.G., S.J., O.P., I.R., L.Z. CROATIA-Vis: H.C., C.H., O.P., I.R., A.F.W. ECRHS: D.L.J., E.O., I.P., M.W. EPIC: N.J.W. FHS: J.B.W., G.T.O. FTC: J.K., K.H.P., T. Rantanen. Health ABC: P.A.C., B.M., W.T. Health 2000: M.H., M.K. KORA F4: S.K., H.S. KORA S3: N.P.-H. NFBC1966: P.E., A.-L.H., M.-R.J., A.P. ORCADES: H.C., S.H.W., J.F.W. RS: G.G.B., M.E., D.W.L., B.H.Ch.S. SHIP: S.G., B.K., H.V. TwinsUK: C.J.H., P.G. Hysi, M.M., T.D.S., G.Z. Stage 2 follow up, ADONIX: J. Brisman, A.-C.O. BHS2: J. Beilby, M.L.H. BRHS: R.W.M., S.G.W., P.H.W. BWHHS: G.D.S., S.E., D.A.L., P.H.W. CARDIA: O.D.W. CROATIA-Split: M.B., I.K., T.Z. GS: SFHS: C.M.J., A.D.M. HCS: C.C., K.A.J., A.A.S. LBC1936: I.J.D., L.M.L., J.M.S. LifeLines: D.S.P., J.M.V. MESA-Lung: R.G.B., J.L.H. Nottingham smokers: K.A.A.B., J.D.B., I.P.H., A. Henry, M.O., I. Sayers. NSHD: R.H., D.K. SAPALDIA: N.P.-H. Look-up studies, ALSPAC: R.G., J. Henderson. ILCCO: ILCCO. Raine: W.Q.A., P.G. Holt, C.E.P., P.D.S.
Genotyping. Stage 1 GWAS, B58C-T1DGC: W.L.M. B58C-WTCCC: W.L.M. BHS1: A.L.J., A.W.M., L.J.P. CHS: S.R.H., B.M.P., J.I.R. CROATIA-Vis: C.H., I.R., A.F.W. ECRHS: M.W. EPIC: I.B., R.J.F.L., J.H.Z. FTC: J.K. Health ABC: Y.L., K.L. Health 2000: S.R., I. Surakka. KORA F4: N.K. KORA S3: C.G. NFBC1966: P.E., A.-L.H., M.-R.J., A.P., A.R. ORCADES: H.C., J.F.W. RS: F.R., A.G.U. SHIP: G.H. TwinsUK: C.J.H., S.-Y.S. Stage 2 follow up, ADONIX: S.D., F.N., A.-C.O. BHS2: J. Beilby, G.C., J.H. BRHS: A.D.H., R.W.M. BWHHS: S.E., D.A.L. CARDIA: M.F., X.G. CROATIA-Split: V.B., T.Z. Gedling: J.R.B., T.M. GS: SFHS: C.M.J., S.M.K., D.J.P. HCS: J.W.H. LBC1936: I.J.D., S.E.H., L.M.L., J.M.S. LifeLines: C.W. MESA-Lung: S.S.R. NSHD: D.K., A.W. SAPALDIA: M.I., F.K. Look-up studies, ALSPAC: S.M.R., W.L.M. ILCCO: ILCCO. Raine: W.Q.A., C.E.P.
Data analysis. Stage 1 GWAS, AGES: G.K.G., A.V.S. ARIC: N.F., D.B.H., L.R.L. B58C–T1DGC: A.R.R., D.P.S. B58C -WTCCC: A.R.R., D.P.S. BHS1: N.M.W. CHS: K.D.M., J.I.R. CROATIA-Korcula: C.H., J.E.H., V.V. CROATIA-Vis: C.H., V.V. ECRHS: D.L.J., A.R. EPIC: J.H.Z. FHS: J.B.W. FTC: I. Surakka. Health ABC: P.A.C., Y.L., K.L., W.T. Health 2000: M.K., S.R., I. Surakka. KORA S3: E.A. NFBC1966: A.R. ORCADES: C.H., V.V. RS: M.E., D.W.L. SHIP: S.G., G.H., B.K., H.V. TwinsUK: M.M., G.Z. Stage 2 follow-up studies, ADONIX: S.D., F.N. BHS2: G.C. BRHS: R.W.M. BWHHS: D.A.L. CARDIA: M.F., X.G. HCS: J.W.H., K.A.J. LBC1936: L.M.L. LifeLines: H.M.B. MESA-Lung: A.M., S.S.R. Nottingham smokers: I. Sayers, A. Henry. NSHD: D.G., R.H. SAPALDIA: I.C., M.I. Look-up studies, ALSPAC: D.M.E. ILCCO: ILCCO. Ox-GSK: J.Z.L. Raine: W.Q.A.
Analysis group: SpiroMeta consortium: I.P.H., T.J., M.S.A., M.D.T., L.V.W. CHARGE consortium: N.F., S.J.L., D.W.L., K.D.M., A.V.S., W.T., J.B.W.
Expression profiling and bioinformatics group: SpiroMeta consortium: I.P.H., M.O., I. Sayers, M.S.A., M.D.T., L.V.W. CHARGE consortium: S.A.G., D.W.L.
Writing group: SpiroMeta consortium: P.E., I.P.H., M.O., M.S.A., D.P.S., M.D.T., L.V.W. CHARGE consortium: S.J.L., D.W.L., S.A.G., G.T.O., V.G., B.H.Ch.S., W.T.
Corresponding authors
Ethics declarations
Competing interests
I.B. and spouse own stock in Incyte Ltd and GlaxoSmithKline. F.N. is employed by AstraZeneca R&D, 431 83 Mölndal, Sweden. D.S.P. has received unrestricted research grants from and has been a consultant to AstraZeneca, Boehringer Ingelheim, Chiesi, GlaxoSmithKline, Nycomed and TEVA. C.F. is a full-time employee of GlaxoSmithKline (GSK), and GSK also funded several aspects of the study as detailed in the Acknowledgements section for Ox-GSK.
Additional information
A full list of members is provided in the Supplementary Note.
A full list of members is provided in the Supplementary Note.
Supplementary information
Supplementary Text and Figures
Supplementary Figure 1, Supplementary Tables 1–6 and Supplementary Note (PDF 3692 kb)
Rights and permissions
About this article
Cite this article
Artigas, M., Loth, D., Wain, L. et al. Genome-wide association and large-scale follow up identifies 16 new loci influencing lung function. Nat Genet 43, 1082–1090 (2011). https://doi.org/10.1038/ng.941
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ng.941