Abstract
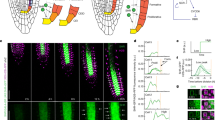
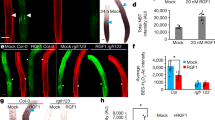
Postmitotic cell growth defines cell shape and size during development. However, the mechanisms regulating postmitotic cell growth in plants remain unknown. Here we report the discovery of a basic helix-loop-helix (bHLH) transcription factor called RSL4 (ROOT HAIR DEFECTIVE 6-LIKE 4) that is sufficient to promote postmitotic cell growth in Arabidopsis thaliana root-hair cells. Loss of RSL4 function resulted in the development of very short root hairs. In contrast, constitutive RSL4 expression programmed constitutive growth, resulting in the formation of very long root hairs. Hair-cell growth signals, such as auxin and low phosphate availability, modulate hair cell extension by regulating RSL4 transcript and protein levels. RSL4 is thus a regulator of growth that integrates endogenous developmental and exogenous environmental signals that together control postmitotic growth in root hairs. The control of postmitotic growth by transcription factors may represent a general mechanism for regulating cell size across diverse organisms.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
Accession codes
References
Schmidt, W. & Schikora, A. Different pathways are involved in phosphate and iron stress-induced alterations of root epidermal cell development. Plant Physiol. 125, 2078–2084 (2001).
Pitts, R.J., Cernac, A. & Estelle, M. Auxin and ethylene promote root hair elongation in Arabidopsis. Plant J. 16, 553–560 (1998).
Bates, T.R. & Lynch, J.P. Stimulation of root hair elongation in Arabidopsis thaliana by low phosphorus availability. Plant Cell Environ. 19, 529–538 (1996).
Menand, B. et al. An ancient mechanism controls the development of cells with a rooting function in land plants. Science 316, 1477–1480 (2007).
Sablowski, R.W.M. & Meyerowitz, E.M. A homolog of NO APICAL MERISTEM is an immediate target of the floral homeotic genes APETALA3/PISTILLATA. Cell 92, 93–103 (1998).
Cho, H.T. & Cosgrove, D.J. Regulation of root hair initiation and expansin gene expression in Arabidopsis. Plant Cell 14, 3237–3253 (2002).
Yao, N., Eisfelder, B.J., Marvin, J. & Greenberg, J.T. The mitochondrion – an organelle commonly involved in programmed cell death in Arabidopsis thaliana. Plant J. 40, 596–610 (2004).
Knox, K., Grierson, C.S. & Leyser, O. AXR3 and SHY2 interact to regulate root hair development. Development 130, 5769–5777 (2003).
Jones, A.R. et al. Auxin transport through non-hair cells sustains root-hair development. Nat. Cell Biol. 11, 78–84 (2009).
Masucci, J.D. & Schiefelbein, J.W. Hormones act downstream of TTG and GL2 to promote root hair outgrowth during epidermis development in the Arabidopsis root. Plant Cell 8, 1505–1517 (1996).
Won, S.-K. et al. Cis-element- and transcriptome-based screening of root hair-specific genes and their functional characterization in Arabidopsis. Plant Physiol. 150, 1459–1473 (2009).
Böhme, K. et al. The Arabidopsis COW1 gene encodes a phosphatidylinositol transfer protein essential for root hair tip growth. Plant J. 40, 686–698 (2004).
Jones, M.A., Raymond, M.J. & Smirnoff, N. Analysis of the root-hair morphogenesis transcriptome reveals the molecular identity of six genes with roles in root-hair development in Arabidopsis. Plant J. 45, 83–100 (2006).
Lasorella, A. et al. Degradation of Id2 by the anaphase-promoting complex couples cell cycle exit and axonal growth. Nature 442, 471–474 (2006).
Johnson, C.M., Stout, P.R., Broyer, T.C. & Carlton, A.B. Comparative chlorine requirements of different plant species. Plant Soil 8, 337–353 (1957).
Ma, Z., Bielenberg, D.G., Brown, K.M. & Lynch, J.P. Regulation of root hair density by phosphorus availability in Arabidopsis thaliana. Plant Cell Environ. 24, 459–467 (2001).
Clough, S.J. & Bent, A.F. Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16, 735–743 (1998).
Shannon, P. et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 13, 2498–2504 (2003).
Maere, S., Heymans, K. & Kuiper, M. BiNGO: a Cytoscape plugin to assess overrepresentation of gene ontology categories in biological networks. Bioinformatics 21, 3448–3449 (2005).
Acknowledgements
L.D. was funded by a grant in aid from the Biotechnology and Biological Sciences Research Council to John Innes Centre and Oxford University. L.D., E.B. and K.Y. were funded by a grant from Human Frontiers in Science (RGP0012/2005-C). B.M. was funded by the EU-Marie Curie Program and a Natural Environmental Research Council (NERC) responsive mode grant NE/C510732/1 to L.D. K.Y. was partially supported by a Joint Scholarship between the University of East Anglia and China Scholarship Council. We gratefully thank the Nottingham Arabidopsis Stock Centre for providing T-DNA insertion and RNAi lines. We are also grateful to Y. Tao for the help with microarray analysis and H. Hu for the statistical analysis.
Author information
Authors and Affiliations
Contributions
K.Y. carried out all experiments except for the identification of genes that act downstream of RSL4, which was carried out by E.B. L.D. and K.Y. wrote the paper with the assistance of B.M. and E.B. K.Y., B.M. and L.D. planned the experiments. L.D. supervised and initiated the study.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Text and Figures
Supplementary Note, Supplementary Figures 1–10 and Supplementary Tables 1 and 2. (PDF 2908 kb)
Rights and permissions
About this article
Cite this article
Yi, K., Menand, B., Bell, E. et al. A basic helix-loop-helix transcription factor controls cell growth and size in root hairs. Nat Genet 42, 264–267 (2010). https://doi.org/10.1038/ng.529
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ng.529
This article is cited by
-
Genome wide association analysis of root hair traits in rice reveals novel genomic regions controlling epidermal cell differentiation
BMC Plant Biology (2023)
-
A mechanical theory of competition between plant root growth and soil pressure reveals a potential mechanism of root penetration
Scientific Reports (2023)
-
OsUGE1 is directly targeted by OsGRF6 to regulate root hair length in rice
Theoretical and Applied Genetics (2023)
-
The Gastrodia menghaiensis (Orchidaceae) genome provides new insights of orchid mycorrhizal interactions
BMC Plant Biology (2022)
-
Two zinc finger proteins with functions in m6A writing interact with HAKAI
Nature Communications (2022)