Abstract
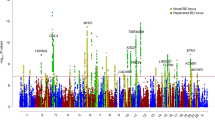
The major mood disorders, which include bipolar disorder and major depressive disorder (MDD), are considered heritable traits, although previous genetic association studies have had limited success in robustly identifying risk loci. We performed a meta-analysis of five case-control cohorts for major mood disorder, including over 13,600 individuals genotyped on high-density SNP arrays. We identified SNPs at 3p21.1 associated with major mood disorders (rs2251219, P = 3.63 × 10−8; odds ratio = 0.87; 95% confidence interval, 0.83–0.92), with supportive evidence for association observed in two out of three independent replication cohorts. These results provide an example of a shared genetic susceptibility locus for bipolar disorder and MDD.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout

Similar content being viewed by others
Accession codes
References
Ustün, T.B., Ayuso-Mateos, J.L., Chatterji, S., Mathers, C. & Murray, C.J. Global burden of depressive disorders in the year 2000. Br. J. Psychiatry 184, 386–392 (2004).
Goodwin, F.K. & Jamison, K. Manic-Depressive Illness 8–26 (Oxford University Press, New York, 2007).
Muglia, P. et al. Genome-wide association study of recurrent major depressive disorder in two European case-control cohorts. Mol. Psychiatry published online, doi: 10.1038/mp.2008.131 (23 December 2008).
Dudbridge, F. & Gusnanto, A. Estimation of significance thresholds for genomewide association scans. Genet. Epidemiol. 32, 227–234 (2008).
Huang, L. et al. Genotype-imputation accuracy across worldwide human populations. Am. J. Hum. Genet. 84, 235–250 (2009).
Scott, L.J. et al. Genome-wide association and meta-analysis of bipolar disorder in individuals of European ancestry. Proc. Natl. Acad. Sci. USA 106, 7501–7506 (2009).
Thompson, M. Polybromo-1: the chromatin targeting subunit of the PBAF complex. Biochimie 91, 309–319 (2009).
Tsai, R.Y.L. & McKay, R.D.G. A nucleolar mechanism controlling cell proliferation in stem cells and cancer cells. Genes Dev. 16, 2991–3003 (2002).
Myers, A.J. et al. A survey of genetic human cortical gene expression. Nat. Genet. 39, 1494–1499 (2007).
Heinzen, E.L. et al. Tissue-specific genetic control of splicing: implications for the study of complex traits. PLoS Biol. 6, e1 (2008).
Doss, S., Schadt, E.E., Drake, T.A. & Lusis, A.J. Cis-acting expression quantitative trait loci in mice. Genome Res. 15, 681–691 (2005).
Moskvina, V. et al. Gene-wide analyses of genome-wide association data sets: evidence for multiple common risk alleles for schizophrenia and bipolar disorder and for overlap in genetic risk. Mol. Psychiatry 14, 252–260 (2009).
Psychiatric GWAS Consortium. A framework for interpreting genome-wide association studies of psychiatric disorders. Mol. Psychiatry 14, 10–17 (2009).
Baum, A.E. et al. A genome-wide association study implicates diacylglycerol kinase eta (DGKH) and several other genes in the etiology of bipolar disorder. Mol. Psychiatry 13, 197–207 (2008).
International Schizophrenia Consortium. Common polygenic variation contributes to risk of schizophrenia and bipolar disorder. Nature 460, 748–752 (2009).
Smith, E.N. et al. Genome-wide association study of bipolar disorder in European American and African American individuals. Mol. Psychiatry 14, 755–763 (2009).
Boomsma, D.I. et al. Genome-wide association of major depression: description of samples for the GAIN Major Depressive Disorder Study: NTR and NESDA biobank projects. Eur. J. Hum. Genet. 16, 335–342 (2008).
Wellcome Trust Case Control Consortium. Genome-wide association study of 14,000 cases of seven common diseases and 3,000 shared controls. Nature 447, 661–678 (2007).
Sklar, P. et al. Whole-genome association study of bipolar disorder. Mol. Psychiatry 13, 558–569 (2008).
Li, Y. & Abecasis, G.R. MACH 1.0: rapid haplotype reconstruction and missing genotype inference. Am. J. Hum. Genet. S79, 2290 (2006).
Purcell, S. et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet. 81, 559–575 (2007).
Devlin, B. & Roeder, K. Genomic control for association studies. Biometrics 55, 997–1004 (1999).
Ioannidis, J.P. Interpretation of tests of heterogeneity and bias in meta-analysis. J. Eval. Clin. Pract. 14, 951–957 (2008).
Sagoo, G.S. et al. Seven lipoprotein lipase gene polymorphisms, lipid fractions, and coronary disease: a HuGE association review and meta-analysis. Am. J. Epidemiol. 168, 1233–1246 (2008).
Littell, J.H., Corcoran, J. & Pillai, V.K. Systematic Reviews and Meta-analysis 62 (Oxford University Press, New York, 2008).
Manolio, T.A., Brooks, L.D. & Collins, F.S.A. HapMap harvest of insights into the genetics of common disease. J. Clin. Invest. 118, 1590–1605 (2008).
Purcell, S., Cherny, S.S. & Sham, P.C. Genetic Power Calculator: design of linkage and association genetic mapping studies of complex traits. Bioinformatics 19, 149–150 (2003).
Pfaffl, M.W. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 29, e45 (2001).
Acknowledgements
Genotyping of the GAIN major depression and NIMH bipolar disorder samples was provided through the Genetic Association Information Network (GAIN), Foundation for NIH (the US National Institutes of Health). The data sets used for the analyses described in this manuscript were obtained from the Database of Genotypes and Phenotypes (dbGaP). Samples and associated phenotype data were provided by the contributing studies. We thank the Wellcome Trust Case Control Consortium, the STEP-BD group, the Netherlands Study of Depression and Anxiety and the Netherlands Twin Registry for making data or results available for analysis. This study used the high-performance computational capabilities of the Biowulf Linux cluster at the NIH. Postmortem brain tissue was supplied by the Stanley Medical Research Institute. Additional acknowledgments are included in the Supplementary Note. Funded by the US NIMH Intramural Research Program, Deutsche Forschungsgemeinschaft (DFG), the National Genome Research Network (NGFN) of the Federal German Ministry of Education and Research, NARSAD (Independent Investigator Award to F.J.M. and Young Investigator Award to T.G.S.), the National German Genome Research Network Plus (NGFNplus) and the MooDS-Net (grant 01GS08144 to S.C. and M.M.N., grant 01GS08147 to M.R.) of the Federal German Ministry of Education and Research, the Heinz Nixdorf Foundation (G. Schmidt, chairman), the Alfried Krupp von Bohlen und Halbach-Stiftung, the DFG Graduate College 793, University of Heidelberg and grants from the NIMH and US National Human Genome Research Institute to J.R.K. (MH078151, MH081804, MH059567 supplement). This research was also supported in part by the Intramural Research Program of the National Library of Medicine, NIH. The replication samples were supported by the Swiss National Science Foundation (3200B0–105993, 32003B-118308 and 33CSCO-122661) and GlaxoSmithKline (Psychiatry Center of Excellence for Drug Discovery and Genetics Division, Verona).
Author information
Authors and Affiliations
Consortia
Contributions
F.J.M. designed the study, led the analysis and wrote the manuscript. N.A., T.G.S. and C.J.M.S. contributed to the data management and analysis. S.D.D.-W., T.G.S., P.M., W.M., F.H., M.R., J.I.N. and H.J.E. (the latter two are members of the BiGS Consortium) edited the manuscript. J.R.W. performed the gene expression experiments. P.M., F.T., R.B., J.S., M.M., T.W.M., W.M, M.M.N., S.C., A.F., J.B.V., F.H., M.P., M.R. and BiGS Consortium members collected samples and/or shared genetic association results. All authors reviewed the manuscript.
Corresponding author
Additional information
A full membership list is provided in the Supplementary Note.
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–2, Supplementary Tables 2–5 and Supplementary Note (PDF 597 kb)
Supplementary Table 1
Complete results of meta-analysis for 317,889 SNPs in five study samples. (XLS 13555 kb)
Rights and permissions
About this article
Cite this article
the Bipolar Disorder Genome Study (BiGS) Consortium. Meta-analysis of genome-wide association data identifies a risk locus for major mood disorders on 3p21.1. Nat Genet 42, 128–131 (2010). https://doi.org/10.1038/ng.523
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ng.523
This article is cited by
-
CADPS functional mutations in patients with bipolar disorder increase the sensitivity to stress
Molecular Psychiatry (2022)
-
Predicting the risk and timing of major mood disorder in offspring of bipolar parents: exploring the utility of a neural network approach
International Journal of Bipolar Disorders (2021)
-
New and sex-specific migraine susceptibility loci identified from a multiethnic genome-wide meta-analysis
Communications Biology (2021)
-
Novel genetic susceptibility loci identified by family based whole exome sequencing in Han Chinese schizophrenia patients
Translational Psychiatry (2020)
-
The genome-wide risk alleles for psychiatric disorders at 3p21.1 show convergent effects on mRNA expression, cognitive function, and mushroom dendritic spine
Molecular Psychiatry (2020)