Abstract
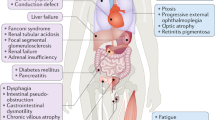
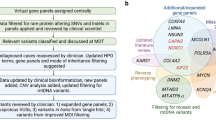
Discovering the molecular basis of mitochondrial respiratory chain disease is challenging given the large number of both mitochondrial and nuclear genes that are involved. We report a strategy of focused candidate gene prediction, high-throughput sequencing and experimental validation to uncover the molecular basis of mitochondrial complex I disorders. We created seven pools of DNA from a cohort of 103 cases and 42 healthy controls and then performed deep sequencing of 103 candidate genes to identify 151 rare variants that were predicted to affect protein function. We established genetic diagnoses in 13 of 60 previously unsolved cases using confirmatory experiments, including cDNA complementation to show that mutations in NUBPL and FOXRED1 can cause complex I deficiency. Our study illustrates how large-scale sequencing, coupled with functional prediction and experimental validation, can be used to identify causal mutations in individual cases.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
References
Skladal, D., Halliday, J. & Thorburn, D.R. Minimum birth prevalence of mitochondrial respiratory chain disorders in children. Brain 126, 1905–1912 (2003).
Distelmaier, F. et al. Mitochondrial complex I deficiency: from organelle dysfunction to clinical disease. Brain 132, 833–842 (2009).
Janssen, R.J., Nijtmans, L.G., van den Heuvel, L.P. & Smeitink, J.A. Mitochondrial complex I: structure, function and pathology. J. Inherit. Metab. Dis. 29, 499–515 (2006).
Lazarou, M., Thorburn, D.R., Ryan, M.T. & McKenzie, M. Assembly of mitochondrial complex I and defects in disease. Biochim. Biophys. Acta 1793, 78–88 (2009).
Bernier, F.P. et al. Diagnostic criteria for respiratory chain disorders in adults and children. Neurology 59, 1406–1411 (2002).
Morava, E. et al. Mitochondrial disease criteria: diagnostic applications in children. Neurology 67, 1823–1826 (2006).
McFarland, R. et al. De novo mutations in the mitochondrial ND3 gene as a cause of infantile mitochondrial encephalopathy and complex I deficiency. Ann. Neurol. 55, 58–64 (2004).
Dimauro, S. & Davidzon, G. Mitochondrial DNA and disease. Ann. Med. 37, 222–232 (2005).
Fontanesi, F., Soto, I.C., Horn, D. & Barrientos, A. Assembly of mitochondrial cytochrome c-oxidase, a complicated and highly regulated cellular process. Am. J. Physiol. Cell Physiol. 291, C1129–C1147 (2006).
Bénit, P. et al. Genotyping microsatellite DNA markers at putative disease loci in inbred/multiplex families with respiratory chain complex I deficiency allows rapid identification of a novel nonsense mutation (IVS1nt −1) in the NDUFS4 gene in Leigh syndrome. Hum. Genet. 112, 563–566 (2003).
Bugiani, M. et al. Clinical and molecular findings in children with complex I deficiency. Biochim. Biophys. Acta 1659, 136–147 (2004).
Lebon, S. et al. Recurrent de novo mitochondrial DNA mutations in respiratory chain deficiency. J. Med. Genet. 40, 896–899 (2003).
Smeitink, J., Sengers, R., Trijbels, F. & van den Heuvel, L. Human NADH:ubiquinone oxidoreductase. J. Bioenerg. Biomembr. 33, 259–266 (2001).
Pagliarini, D.J. et al. A mitochondrial protein compendium elucidates complex I disease biology. Cell 134, 112–123 (2008).
Marcotte, E.M., Pellegrini, M., Thompson, M.J., Yeates, T.O. & Eisenberg, D. A combined algorithm for genome-wide prediction of protein function. Nature 402, 83–86 (1999).
Pellegrini, M., Marcotte, E.M., Thompson, M.J., Eisenberg, D. & Yeates, T.O. Assigning protein functions by comparative genome analysis: protein phylogenetic profiles. Proc. Natl. Acad. Sci. USA 96, 4285–4288 (1999).
Ogilvie, I., Kennaway, N.G. & Shoubridge, E.A. A molecular chaperone for mitochondrial complex I assembly is mutated in a progressive encephalopathy. J. Clin. Invest. 115, 2784–2792 (2005).
Saada, A. et al. Mutations in NDUFAF3 (C3ORF60), encoding an NDUFAF4 (C6ORF66)-interacting complex I assembly protein, cause fatal neonatal mitochondrial disease. Am. J. Hum. Genet. 84, 718–727 (2009).
Sugiana, C. et al. Mutation of C20orf7 disrupts complex I assembly and causes lethal neonatal mitochondrial disease. Am. J. Hum. Genet. 83, 468–478 (2008).
Bentley, D.R. et al. Accurate whole human genome sequencing using reversible terminator chemistry. Nature 456, 53–59 (2008).
Frazer, K.A. et al. A second generation human haplotype map of over 3.1 million SNPs. Nature 449, 851–861 (2007).
Sherry, S.T. et al. dbSNP: the NCBI database of genetic variation. Nucleic Acids Res. 29, 308–311 (2001).
Ingman, M. & Gyllensten, U. mtDB: Human Mitochondrial Genome Database, a resource for population genetics and medical sciences. Nucleic Acids Res. 34, D749–D751 (2006).
Stenson, P.D. et al. The Human Gene Mutation Database: 2008 update. Genome Med 1, 13 (2009).
Kirby, D.M. et al. NDUFS6 mutations are a novel cause of lethal neonatal mitochondrial complex I deficiency. J. Clin. Invest. 114, 837–845 (2004).
Valente, L. et al. Identification of novel mutations in five patients with mitochondrial encephalomyopathy. Biochim. Biophys. Acta 1787, 491–501 (2009).
Budde, S.M. et al. Combined enzymatic complex I and III deficiency associated with mutations in the nuclear encoded NDUFS4 gene. Biochem. Biophys. Res. Commun. 275, 63–68 (2000).
Leshinsky-Silver, E. et al. NDUFS4 mutations cause Leigh syndrome with predominant brainstem involvement. Mol. Genet. Metab. 97, 185–189 (2009).
Petruzzella, V. et al. A nonsense mutation in the NDUFS4 gene encoding the 18 kDa (AQDQ) subunit of complex I abolishes assembly and activity of the complex in a patient with Leigh-like syndrome. Hum. Mol. Genet. 10, 529–535 (2001).
Anderson, S.L. et al. A novel mutation in NDUFS4 causes Leigh syndrome in an Ashkenazi Jewish family. J. Inherit. Metab. Dis. 32, 121 (2009).
van den Heuvel, L. et al. Demonstration of a new pathogenic mutation in human complex I deficiency: a 5-bp duplication in the nuclear gene encoding the 18-kD (AQDQ) subunit. Am. J. Hum. Genet. 62, 262–268 (1998).
Schuelke, M. et al. Mutant NDUFV1 subunit of mitochondrial complex I causes leukodystrophy and myoclonic epilepsy. Nat. Genet. 21, 260–261 (1999).
Loeffen, J. et al. The first nuclear-encoded complex I mutation in a patient with Leigh syndrome. Am. J. Hum. Genet. 63, 1598–1608 (1998).
McKenzie, M., Lazarou, M., Thorburn, D.R. & Ryan, M.T. Mitochondrial respiratory chain supercomplexes are destabilized in Barth Syndrome patients. J. Mol. Biol. 361, 462–469 (2006).
Choi, M. et al. Genetic diagnosis by whole exome capture and massively parallel DNA sequencing. Proc. Natl. Acad. Sci. USA 106, 19096–19101 (2009).
Ng, S.B. et al. Targeted capture and massively parallel sequencing of 12 human exomes. Nature 461, 272–276 (2009).
Ng, S.B. et al. Exome sequencing identifies the cause of a mendelian disorder. Nat. Genet. 42, 30–35 (2010).
Sheftel, A.D. et al. Human ind1, an iron-sulfur cluster assembly factor for respiratory complex I. Mol. Cell. Biol. 29, 6059–6073 (2009).
Bych, K. et al. The iron-sulphur protein Ind1 is required for effective complex I assembly. EMBO J. 27, 1736–1746 (2008).
Calvo, S. et al. Systematic identification of human mitochondrial disease genes through integrative genomics. Nat. Genet. 38, 576–582 (2006).
Tarpey, P.S. et al. A systematic, large-scale resequencing screen of X-chromosome coding exons in mental retardation. Nat. Genet. 41, 535–543 (2009).
Kirby, D.M. et al. Respiratory chain complex I deficiency: an underdiagnosed energy generation disorder. Neurology 52, 1255–1264 (1999).
Gnirke, A. et al. Solution hybrid selection with ultra-long oligonucleotides for massively parallel targeted sequencing. Nat. Biotechnol. 27, 182–189 (2009).
Li, H., Ruan, J. & Durbin, R. Mapping short DNA sequencing reads and calling variants using mapping quality scores. Genome Res. 18, 1851–1858 (2008).
Li, H. et al. The Sequence Alignment/Map format and SAMtools. Bioinformatics 25, 2078–2079 (2009).
Calvo, S.E., Pagliarini, D.J. & Mootha, V.K. Upstream open reading frames cause widespread reduction of protein expression and are polymorphic among humans. Proc. Natl. Acad. Sci. USA 106, 7507–7512 (2009).
Karolchik, D., Hinrichs, A.S. & Kent, W.J. The UCSC Genome Browser. Curr. Protoc. Bioinformatics Chapter 1: Unit 1.4 (2009).
Dimmic, M.W., Sunyaev, S. & Bustamante, C.D. Inferring SNP function using evolutionary, structural, and computational methods. Pac. Symp. Biocomput. 382–384 (2005).
Cree, L.M., Samuels, D.C. & Chinnery, P.F. The inheritance of pathogenic mitochondrial DNA mutations. Biochim. Biophys. Acta 1792, 1097–1102 (2009).
Gabriel, S., Ziaugra, L. & Tabbaa, D. SNP genotyping using the Sequenom MassARRAY iPLEX platform. Curr. Protoc. Hum. Genet. Chapter 2: Unit 2.12 (2009).
Lamandé, S.R. et al. Reduced collagen VI causes Bethlem myopathy: a heterozygous COL6A1 nonsense mutation results in mRNA decay and functional haploinsufficiency. Hum. Mol. Genet. 7, 981–989 (1998).
Acknowledgements
We thank S. Tregoning, A. Laskowski and S. Smith for assistance with enzyme assays and DNA preparation, M. McKenzie and M. Ryan for the NDUFAF2 antibody, J. Boehm for the lentiviral expression vector, S. Flynn for assistance with human subjects protocols, R. Onofrio for designing PCR primers, K. Ardlie and S. Mahan for assistance in DNA sample preparation, J. Wilkinson and L. Ambrogio for Illumina sequence project management, T. Fennel for sequence alignment, L. Ziaugra for genotyping assistance, M. Cabili for tool evaluation, J. Flannick for assistance with pooled sequence analysis, I. Adzhubei and S. Sunyaev for PolyPhen-2.0 predictions, M. DePristo, E. Banks and A. Sivachenko for advice on sequence data analysis, M. Garber for assistance with evolutionary conservation analyses, J. Pirruccello, R. Do and S. Kathiresan for data and analysis of control data, and the many physicians who referred subjects and assisted with these studies. This work was supported by a grant (436901) and Principal Research Fellowship from the Australian National Health & Medical Research Council awarded to D.R.T., an Australian Postgraduate Award to E.J.T. and a grant from the US National Institutes of Health (GM077465) to V.K.M. The authors wish to dedicate this article to the memory of our co-author Denise Kirby, an outstanding scientist and dear colleague who died during the preparation of this manuscript.
Author information
Authors and Affiliations
Contributions
This study was conceived and designed by S.E.C., D.R.T. and V.K.M. with input from M.J.D. and S.B.G. Enzyme diagnosis of the cohort was coordinated by D.M.K. E.W. and C.J.W. provided clinical interaction and assisted with sample collection. Samples were collected by D.M.K., E.W. and C.J.W. and prepared by A.G.C. and E.J.T. The pooled sequencing protocol was designed and established at the Broad Institute by D.A., M.J.D. and S.B.G. Project management was performed by S.E.C., N.P.B. and C.G. G.C. performed pooling. M.C.R. and C.G. performed the genotyping. S.E.C. designed and performed the computational analyses, with assistance from E.J.T., A.G.C. and M.R. All experiments were designed and performed by E.J.T., A.G.C. and O.A.G. Affymetrix array-based cytogenetic analysis was performed by D.L.B. Syzygy was developed and run by M.R. and M.J.D. The manuscript was written by S.E.C., E.J.T., A.G.C., D.R.T. and V.K.M. All aspects of the study were supervised by D.R.T. and V.K.M.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–10, Supplementary Tables 1, 3–5 and Supplementary Note (PDF 853 kb)
Supplementary Table 2
Likely deleterious variants detected and validated in 103 patients with Complex I deficiency (XLS 103 kb)
Rights and permissions
About this article
Cite this article
Calvo, S., Tucker, E., Compton, A. et al. High-throughput, pooled sequencing identifies mutations in NUBPL and FOXRED1 in human complex I deficiency. Nat Genet 42, 851–858 (2010). https://doi.org/10.1038/ng.659
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ng.659
This article is cited by
-
Mitochondrial proteome research: the road ahead
Nature Reviews Molecular Cell Biology (2024)
-
Deficiency of the mitochondrial ribosomal subunit, MRPL50, causes autosomal recessive syndromic premature ovarian insufficiency
Human Genetics (2023)
-
TEFM variants impair mitochondrial transcription causing childhood-onset neurological disease
Nature Communications (2023)
-
Early embryonic lethality in complex I associated p.L104P Nubpl mutant mice
Orphanet Journal of Rare Diseases (2022)
-
Compound heterozygous mutations of NDUFV1 identified in a child with mitochondrial complex I deficiency
Genes & Genomics (2022)