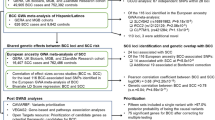
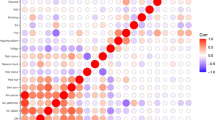
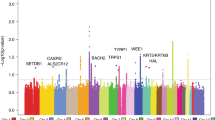
Abstract
In a follow-up to our previously reported genome-wide association study of cutaneous basal cell carcinoma (BCC)1, we describe here several new susceptibility variants. SNP rs11170164, encoding a G138E substitution in the keratin 5 (KRT5) gene, affects risk of BCC (OR = 1.35, P = 2.1 × 10−9). A variant at 9p21 near CDKN2A and CDKN2B also confers susceptibility to BCC (rs2151280[C]; OR = 1.19, P = 6.9 × 10−9), as does rs157935[T] at 7q32 near the imprinted gene KLF14 (OR = 1.23, P = 5.7 × 10−10). The effect of rs157935[T] is dependent on the parental origin of the risk allele. None of these variants were found to be associated with melanoma or fair-pigmentation traits. A melanoma- and pigmentation-associated variant in the SLC45A2 gene, L374F, is associated with risk of both BCC and squamous cell carcinoma. Finally, we report conclusive evidence that rs401681[C] in the TERT-CLPTM1L locus confers susceptibility to BCC but protects against melanoma.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout

Similar content being viewed by others
References
Stacey, S.N. et al. Common variants on 1p36 and 1q42 are associated with cutaneous basal cell carcinoma but not with melanoma or pigmentation traits. Nat. Genet. 40, 1313–1318 (2008).
Epstein, E.H. Basal cell carcinomas: attack of the hedgehog. Nat. Rev. Cancer 8, 743–754 (2008).
Gudbjartsson, D.F. et al. ASIP and TYR pigmentation variants associate with cutaneous melanoma and basal cell carcinoma. Nat. Genet. 40, 886–891 (2008).
Rafnar, T. et al. Sequence variants at the TERT-CLPTM1L locus associate with many cancer types. Nat. Genet. 41, 221–227 (2009).
Guedj, M. et al. Variants of the MATP/SLC45A2 gene are protective for melanoma in the French population. Hum. Mutat. 29, 1154–1160 (2008).
Fernandez, L.P. et al. SLC45A2: a novel malignant melanoma-associated gene. Hum. Mutat. 29, 1161–1167 (2008).
Moll, R., Divo, M. & Langbein, L. The human keratins: biology and pathology. Histochem. Cell Biol. 129, 705–733 (2008).
Sulem, P. et al. Two newly identified genetic determinants of pigmentation in Europeans. Nat. Genet. 40, 835–837 (2008).
Sulem, P. et al. Genetic determinants of hair, eye and skin pigmentation in Europeans. Nat. Genet. 39, 1443–1452 (2007).
Uitto, J., Richard, G. & McGrath, J.A. Diseases of epidermal keratins and their linker proteins. Exp. Cell Res. 313, 1995–2009 (2007).
Chan, Y.M., Yu, Q.C., Fine, J.D. & Fuchs, E. The genetic basis of Weber-Cockayne epidermolysis bullosa simplex. Proc. Natl. Acad. Sci. USA 90, 7414–7418 (1993).
Kowalewski, C. et al. A novel autosomal partially dominant mutation designated G476D in the keratin 5 gene causing epidermolysis bullosa simplex Weber-Cockayne type: a family study with a genetic twist. Int. J. Mol. Med. 20, 75–78 (2007).
Shurman, D. et al. Epidermolysis Bullosa Simplex with mottled pigmentation: mutation analysis in the first reported Hispanic pedigree with the largest single generation of affected individuals to date. Eur. J. Dermatol. 16, 132–135 (2006).
Kim, W.Y. & Sharpless, N.E. The regulation of INK4/ARF in cancer and aging. Cell 127, 265–275 (2006).
Pasmant, E. et al. Characterization of a germ-line deletion, including the entire INK4/ARF locus, in a melanoma-neural system tumor family: identification of ANRIL, an antisense noncoding RNA whose expression coclusters with ARF. Cancer Res. 67, 3963–3969 (2007).
Lin, J., Hocker, T.L., Singh, M. & Tsao, H. Genetics of melanoma predisposition. Br. J. Dermatol. 159, 286–291 (2008).
Helgadottir, A. et al. A common variant on chromosome 9p21 affects the risk of myocardial infarction. Science 316, 1491–1493 (2007).
Saxena, R. et al. Genome-wide association analysis identifies loci for type 2 diabetes and triglyceride levels. Science 316, 1331–1336 (2007).
Scott, L.J. et al. A genome-wide association study of type 2 diabetes in Finns detects multiple susceptibility variants. Science 316, 1341–1345 (2007).
Zeggini, E. et al. Replication of genome-wide association signals in UK samples reveals risk loci for type 2 diabetes. Science 316, 1336–1341 (2007).
Steinthorsdottir, V. et al. A variant in CDKAL1 influences insulin response and risk of type 2 diabetes. Nat. Genet. 39, 770–775 (2007).
Mishmar, D. et al. Molecular characterization of a common fragile site (FRA7H) on human chromosome 7 by the cloning of a simian virus 40 integration site. Proc. Natl. Acad. Sci. USA 95, 8141–8146 (1998).
Parker-Katiraee, L. et al. Identification of the imprinted KLF14 transcription factor undergoing human-specific accelerated evolution. PLoS Genet. 3, e65 (2007).
Kong, A. et al. Detection of sharing by descent, long-range phasing and haplotype imputation. Nat. Genet. 40, 1068–1075 (2008).
Fabbri, M. et al. MicroRNA-29 family reverts aberrant methylation in lung cancer by targeting DNA methyltransferases 3A and 3B. Proc. Natl. Acad. Sci. USA 104, 15805–15810 (2007).
Graf, J., Hodgson, R. & van Daal, A. Single nucleotide polymorphisms in the MATP gene are associated with normal human pigmentation variation. Hum. Mutat. 25, 278–284 (2005).
Han, J. et al. A prospective study of telomere length and the risk of skin cancer. J. Invest. Dermatol. 129, 415–421 (2008).
Bataille, V. et al. Nevus size and number are associated with telomere length and represent potential markers of a decreased senescence in vivo. Cancer Epidemiol. Biomarkers Prev. 16, 1499–1502 (2007).
Higgins, J.P. & Thompson, S.G. Quantifying heterogeneity in a meta-analysis. Stat. Med. 21, 1539–1558 (2002).
Szeverenyi, I. et al. The Human Intermediate Filament Database: comprehensive information on a gene family involved in many human diseases. Hum. Mutat. 29, 351–360 (2008).
Acknowledgements
We thank G.H. Olafsdottir and L. Tryggvadottir of the Icelandic Cancer Registry for assistance with the ascertainment of affected individuals, S. Sveinsdottir for assistance with subject recruitment, and H. Sigurdsson for assistance with the figures. We are grateful to I. Saaf for permission to reproduce a section of the Human Intermediate Filament Database in Supplementary Figure 1. This study was supported in part by the Jubilaumsfonds of the Austrian National Bank (project numbers 11946 and 12161), the Swedish Cancer Society, the Radiumhemmet Research Funds and the Swedish Research Council, the US National Institute of Environmental Health Sciences (T32E007155), the US National Institutes of Health (R01CA082354 and R01CA57494), and a Research Investment Grant of the Radboud University Nijmegen Medical Centre.
Author information
Authors and Affiliations
Contributions
The study was designed and results were interpreted by S.N.S., P.S., J.R.G., T.R., A.K., J.H.O., U.T. and K.S. Patient ascertainment and recruitment was organized and carried out by S.N.S., B. Sigurgeirsson, K.R.B., K.T., R.R., D.S., K.H., P.R., E.G., K. Koppova, R.B.-E., V. Soriano, P.J., B. Saez, Y.G., V.F., C.C., M.G., V.H., A.L., J.J.B., M.M.v.R., K.K.H.A., E.d.V., M.S., M.G.D.M., A.M., J.W., P.H., H.P., J.G., S.G., H.H., V. Steinthorsdottir, K. Kristjansson, G.B., I.O., L.R., M.R., L.A.K., J.H., E.N., J.I.M., R.K., M.R.K., H.H.N., J.H.O. and U.T. Biological material collection and handling was supervised by S.N.S., D.S., P.R., E.G., K. Koppova, B. Saez, V.H., A.L., K.K.H.A., J.S., H.B., I.O., M.R., M.R.K., H.H.N. and U.T. Genotyping was supervised by S.N.S., M.J., A.S., J.G., D.N.M., S.G., H.H., V. Steinthorsdottir, T.B., T.R. and U.T. Statistical analysis was carried out by P.S., G.T., D.F.G., M.L.F. and A.K. Bioinformatic analysis was carried out by S.N.S., S.A.G. and G.M. Authors S.N.S., P.S., A.K. and K.S. drafted the manuscript. All authors contributed to the final version of the paper. Principal collaborators for the case-control population samples were J.H.O. (Iceland), H.H.N. and M.R.K. (USA), R.K. (Eastern Europe), J.I.M. and E.N. (Spain), J.H. (Sweden), L.A.K. (The Netherlands), M.R. (Italy) and I.O. (Austria).
Corresponding authors
Ethics declarations
Competing interests
Authors whose affiliations are listed as deCODE Genetics are shareholders and/or employees of deCODE Genetics, a biotechnology company. deCODE Genetics intends to incorporate the variants described in this paper into its genetic testing services.
Supplementary information
Supplementary Text and Figures
Supplementary Tables 1–8 and Supplementary Figure 1 (PDF 1739 kb)
Rights and permissions
About this article
Cite this article
Stacey, S., Sulem, P., Masson, G. et al. New common variants affecting susceptibility to basal cell carcinoma. Nat Genet 41, 909–914 (2009). https://doi.org/10.1038/ng.412
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ng.412
This article is cited by
-
Multi-ancestry genome-wide meta-analysis identifies novel basal cell carcinoma loci and shared genetic effects with squamous cell carcinoma
Communications Biology (2024)
-
KLF14 regulates the growth of hepatocellular carcinoma cells via its modulation of iron homeostasis through the repression of iron-responsive element-binding protein 2
Journal of Experimental & Clinical Cancer Research (2023)
-
A systematic review of skin ageing genes: gene pleiotropy and genes on the chromosomal band 16q24.3 may drive skin ageing
Scientific Reports (2022)
-
Genetic ancestry, skin pigmentation, and the risk of cutaneous squamous cell carcinoma in Hispanic/Latino and non-Hispanic white populations
Communications Biology (2020)
-
Genome-wide meta-analysis identifies eight new susceptibility loci for cutaneous squamous cell carcinoma
Nature Communications (2020)