Abstract
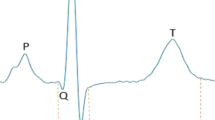
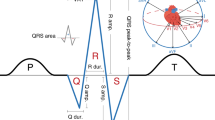
The QT interval, a measure of cardiac repolarization, predisposes to ventricular arrhythmias and sudden cardiac death (SCD) when prolonged or shortened. A common variant in NOS1AP is known to influence repolarization. We analyze genome-wide data from five population-based cohorts (ARIC, KORA, SardiNIA, GenNOVA and HNR) with a total of 15,842 individuals of European ancestry, to confirm the NOS1AP association and identify nine additional loci at P < 5 × 10−8. Four loci map near the monogenic long-QT syndrome genes KCNQ1, KCNH2, SCN5A and KCNJ2. Two other loci include ATP1B1 and PLN, genes with established electrophysiological function, whereas three map to RNF207, near LITAF and within NDRG4-GINS3-SETD6-CNOT1, respectively, all of which have not previously been implicated in cardiac electrophysiology. These results, together with an accompanying paper from the QTGEN consortium, identify new candidate genes for ventricular arrhythmias and SCD.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
References
Arking, D.E., Chugh, S.S., Chakravarti, A. & Spooner, P.M. Genomics in sudden cardiac death. Circ. Res. 94, 712–723 (2004).
Tomaselli, G.F. et al. Sudden cardiac death in heart failure. The role of abnormal repolarization. Circulation 90, 2534–2539 (1994).
Vrtovec, B. et al. Prolonged QTc interval and high B-type natriuretic peptide levels together predict mortality in patients with advanced heart failure. Circulation 107, 1764–1769 (2003).
Schouten, E.G. et al. QT interval prolongation predicts cardiovascular mortality in an apparently healthy population. Circulation 84, 1516–1523 (1991).
Moss, A.J. et al. The long QT syndrome: prospective longitudinal study of 328 families. Circulation 84, 1136–1144 (1991).
Arking, D.E. et al. A common genetic variant in the NOS1 regulator NOS1AP modulates cardiac repolarization. Nat. Genet. 38, 644–651 (2006).
Aarnoudse, A.J. et al. Common NOS1AP variants are associated with a prolonged QTc interval in the Rotterdam Study. Circulation 116, 10–11 (2007).
Post, W. et al. Associations between genetic variants in the NOS1AP (CAPON) gene and cardiac repolarization in the old order Amish. Hum. Hered. 64, 214–219 (2007).
Tobin, M.D. et al. Gender and effects of a common genetic variant in the NOS1 regulator NOS1AP on cardiac repolarization in 3761 individuals from two independent populations. Int. J. Epidemiol. 37, 1132–1141 (2008).
Kao, W.H.L. et al. Genetic variations in NOS1AP are associated with sudden cardiac death in U.S. white community based populations. Circulation 24, 940–951 (2009).
ARIC Investigators. The Atherosclerosis Risk in Communities (ARIC) Study: design and objectives. The ARIC investigators. Am. J. Epidemiol 129, 687–702 (1989).
Pilia, G. et al. Heritability of cardiovascular and personality traits in 6,148 Sardinians. PLoS Genet. 2, e132 (2006).
Wichmann, H.E., Gieger, C. & Illig, T. KORA-gen–resource for population genetics, controls and a broad spectrum of disease phenotypes. Gesundheitswesen 67 (Suppl. 1), S26–S30 (2005).
Pattaro, C. et al. The genetic study of three population microisolates in South Tyrol (MICROS): study design and epidemiological perspectives. BMC Med. Genet. 8, 29 (2007).
Schmermund, A. et al. Assessment of clinically silent atherosclerotic disease and established and novel risk factors for predicting myocardial infarction and cardiac death in healthy middle-aged subjects: rationale and design of the Heinz Nixdorf Recall Study. Am. Heart J. 144, 212–218 (2002).
The International HapMap Consortium. A haplotype map of the human genome. Nature 437, 1299–1320 (2005).
Devlin, B. & Roeder, K. Genomic control for association studies. Biometrics 55, 997–1004 (1999).
Splawski, I. et al. Variant of SCN5A sodium channel implicated in risk of cardiac arrhythmia. Science 297, 1333–1336 (2002).
Bezzina, C.R. et al. A common polymorphism in KCNH2 (HERG) hastens cardiac repolarization. Cardiovasc. Res. 59, 27–36 (2003).
Pfeufer, A. et al. Common variants in myocardial ion channel genes modify the QT interval in the general population: results from the KORA study. Circ. Res. 96, 693–701 (2005).
Newton-Cheh, C. et al. Common genetic variation in KCNH2 is associated with QT interval duration: the Framingham Heart Study. Circulation 116, 1128–1136 (2007).
Gouas, L. et al. D.E.S.I.R. Study Group. Association of KCNQ1, KCNE1, KCNH2 and SCN5A polymorphisms with QTc interval length in a healthy population. Eur. J. Hum. Genet. 13, 1213–1222 (2005).
Emison, E.S. et al. A common, sex-dependent mutation in a putative RET enhancer underlies Hirschsprung disease susceptibility. Nature 434, 857–863 (2005).
Liu, Y. et al. The human inward rectifier K(+) channel subunit kir5.1 (KCNJ16) maps to chromosome 17q25 and is expressed in kidney and pancreas. Cytogenet. Cell Genet. 90, 60–63 (2000).
Chang, Y.P. et al. Multiple genes for essential-hypertension susceptibility on chromosome 1q. Am. J. Hum. Genet. 80, 253–264 (2007).
Schmitt, J.P. et al. Dilated cardiomyopathy and heart failure caused by a mutation in phospholamban. Science 299, 1410–1413 (2003).
Street, V.A. et al. Mutation of a putative protein degradation gene LITAF/SIMPLE in Charcot-Marie-Tooth disease 1C. Neurology 60, 22–26 (2003).
Qu, X. et al. Ndrg4 is required for normal myocyte proliferation during early cardiac development in zebrafish. Dev. Biol. 317, 486–496 (2008).
Scherf, M., Klingenhoff, A. & Werner, T. Highly specific localization of promoter regions in large genomic sequences by PromoterInspector: a novel context analysis approach. J. Mol. Biol. 297, 599–606 (2000).
Hiroi, Y. et al. Tbx5 associates with Nkx2–5 and synergistically promotes cardiomyocyte differentiation. Nat. Genet. 28, 276–280 (2001).
Cohen, J.C. et al. Multiple rare alleles contribute to low plasma levels of HDL cholesterol. Science 305, 869–872 (2004).
Rautaharju, P.M. et al. Sex differences in the evolution of the electrocardiographic QT interval with age. Can. J. Cardiol. 8, 690–695 (1992).
Arking, D.E. et al. Multiple independent genetic factors at NOS1AP modulate the QT interval in a multi-ethnic population. PLoS ONE 4, e4333 (2009).
Newton-Cheh, C. et al. Common variants at ten loci influence QT interval duration in the QTGEN Study. Nat. Genet. advance online publication, doi: 10.1038/ng.361 (22 March 2009).
Friedlander, Y. et al. Possible association of the human KCNE1 (minK) gene and QT interval in healthy subjects: evidence from association and linkage analyses in Israeli families. Ann. Hum. Genet. 69, 645–656 (2005).
Akyol, M. et al. The common non-synonymous variant G38S of the KCNE1-(minK)-gene is not associated to QT interval in Central European Caucasians: results from the KORA study. Eur. Heart J. 28, 305–309 (2007).
Weir, B.S. & Clark Cockerham, C. Estimating F-statistics for the analysis of population structure. Evolution Int. J. Org. Evolution 38, 1358–1370 (1984).
Manolio, T., Brooks, L. & Collins, F.A. HapMap harvest of insights into the genetics of common disease. J. Clin. Invest. 118, 1590–1605 (2008).
Willer, C.J. et al. Newly identified loci that influence lipid concentrations and risk of coronary artery disease. Nat. Genet. 40, 161–169 (2008).
Sanna, S. et al. Common variants in the GDF5-UQCC region are associated with variation in human height. Nat. Genet. 40, 198–203 (2008).
Döring, A. et al. SLC2A9 influences uric acid concentrations with pronounced sex-specific effects. Nat. Genet. 40, 430–436 (2008).
Acknowledgements
We gratefully acknowledge all participants in the community-based studies of ARIC, KORA, SardiNIA, GenNOVA and Heinz Nixdorf Recall Study, and all the members of our laboratories for helpful discussion of this study. We thank G. Fischer for help with genotype imputation and data management for the KORA samples, Y. Li for her help on statistical analysis, K. Tarasov for in silico promotor analysis, A. Cao for his valuable advice and support in the SardiNIA project and all ARIC, KORA, SardiNIA, GenNOVA and Heinz Nixdorf Recall Study investigators for study design and continued operation.
We also thank C. Egger and Y. D'Elia for the valuable support in data management and data administration; S. Melville and M. Facheris for the important work of drug classification in the GenNOVA project; and the primary care practitioners R. Stocker, S. Waldner, T. Pizzecco, J. Plangger, U. Marcadent and the personnel of the Hospital of Silandro (Department of Laboratory Medicine) for their participation and collaboration in the GenNOVA project.
ARIC is carried out as a collaborative study supported by National Heart, Lung, and Blood Institute contracts N01-HC-55015, N01-HC-55016, N01-HC-55018, N01-HC-55019, N01-HC-55020, N01-HC-55021, N01-HC-55022, R01HL087641, R01HL59367 and R01HL086694; National Human Genome Research Institute contract U01HG004402; and National Institutes of Health contract HHSN268200625226C. Infrastructure was partly supported by Grant Number UL1RR025005, a component of the National Institutes of Health and NIH Roadmap for Medical Research. In addition, we acknowledge support from NHLBI grants HL86694 and HL054512, and the Donald W. Reynolds Cardiovascular Clinical Research Center at Johns Hopkins University for genotyping and data analysis relevant to this study. A.K. is supported by a German Research Foundation Fellowship.
The KORA study was funded by grants by the German Federal Ministry of Education and Research (BMBF) in the context of the German National Genome Research Network (NGFN), the German National Competence network on atrial fibrillation (AFNET) and the Bioinformatics for the Functional Analysis of Mammalian Genomes program (BFAM) by grants to S.K. (NGFN 01GS0499, 01GS0838 and AF-Net 01GI0204/N), A.P. (NGFN 01GR0803, 01EZ0874), H.–E.W. (NGFN 01GI0204) and to T.M. (NGFN 01GR0103). S.K. is also supported by a grant from the Fondation Leducq. The KORA platform is funded by the BMBF and by the State of Bavaria.
The SardiNIA team was supported by Contract NO1-AG-1-2109 from the National Institute on Aging and in part by the Intramural Research Program of the US National Institute on Aging, NIH. The efforts of G.R.A. were supported in part by contract 263-MA-410953 from the National Institute on Aging to the University of Michigan and by research grants from the National Human Genome Research Institute and the National Heart, Lung, and Blood Institute (to G.R.A.).
The GenNOVA study was supported by the Ministry of Health of the Autonomous Province of Bolzano and the South Tyrolean Sparkasse Foundation.
The Heinz Nixdorf Recall Study was funded by a grant of the Heinz Nixdorf Foundation (Chairman: G. Schmidt).
Author information
Authors and Affiliations
Contributions
Designed the scientific rationale of the work: A.P., S.S., D.E.A., R.J.P., S.S.N., A.A.H., D.S., T.M., M.U., J.C., S.K., G.R.A., A.C. Obtained funding: A.P., R.E., K.-H.J., S.N., G.S., A.A.H., E.L., H.-E.W., D.S., E.B., T.M., M.U., J.C., S.K., G.R.A., A.C. Analyzed EKG readings: A.P., S.S., D.E.A., C.F., M.O., S.P., M.F.S., B.M.-M., C.H., S.S.N., S.M., F.M., E.L. Performed genotyping: A.P., S.S., D.E.A., T.W.M., A.A.H., P.P.P., T.M., A.C. Performed genotype imputation: A.P., S.S., D.E.A., M.M., C.P., B.P., T.W.M., B.M.-M., A.A.H. Performed statistical analysis: A.P., S.S., D.E.A., M.M., C.F., C.P., G.B.E., C.G., F.M. Performed metaanalysis: A.P., S.S., D.E.A., V.G., M.M., C.P. Drafted manuscript: A.P., S.S., D.E.A., G.R.A., A.C. Critically revised manuscript: M.M., M.O., A.K., G.U., M.B., M.L., A.S., M.F.S., C.H., T.W.M., M.D., S.M., L.C., R.E., K.-H.J., S.N., G.S., F.M., A.A.H., B.M.-M., P.P.P., H.-E.W., D.S., E.B., T.M., M.U., J.C.
List of investigators by cohort:
ARIC (Atherosclerosis Risk in Communities Study): D.E.A., G.B.E., A.K., M.B., M.L., R.J.P., W.H.L.K., E.B., J.C., A.C.
GenNOVA: F.M., C.P., C.F., A.A.H., P.P.P.
KORA (Kooperative Gesundheitsforschung in der Region Ausgsburg): A.P., M.M., S.P., M.F.S., C.H., C.G., B.P., B.M.-M., G.S., S.K., H.-E.W., T.M.
HNR (Heinz Nixdorf Recall Study): T.W.M., S.M., R.E., K.-H.J.
SardiNIA: S.S., V.G., M.O., G.U., A.S., S.N., M.D., L.C., S.N., E.L., D.S., M.U., G.R.A.
Corresponding authors
Ethics declarations
Competing interests
Aravinda Chakravarti is on the Scientific Advisory Board of Affymetrix. This potential conflict of interest is managed by the policies of Johns Hopkins University.
Supplementary information
Supplementary Text and Figures
Supplementary Tables 1–8, Supplementary Figure 1 and Supplementary Methods (PDF 1020 kb)
Rights and permissions
About this article
Cite this article
Pfeufer, A., Sanna, S., Arking, D. et al. Common variants at ten loci modulate the QT interval duration in the QTSCD Study. Nat Genet 41, 407–414 (2009). https://doi.org/10.1038/ng.362
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ng.362
This article is cited by
-
Thyroid hormones regulate cardiac repolarization and QT-interval related gene expression in hiPSC cardiomyocytes
Scientific Reports (2022)
-
Evaluating Common NOS1AP Variants in Patients with Implantable Cardioverter Defibrillators for Secondary Prevention
Journal of Interventional Cardiac Electrophysiology (2022)
-
Association of T66A polymorphism in CASQ2 with PR interval in a Chinese population
Herz (2021)
-
Exome sequencing identifies a novel nonsense mutation of Ring Finger Protein 207 in a Chinese family with Long QT syndrome and syncope
Journal of Human Genetics (2019)
-
Nitric oxide signalling in cardiovascular health and disease
Nature Reviews Cardiology (2018)