Abstract
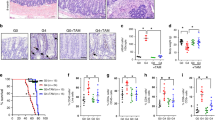
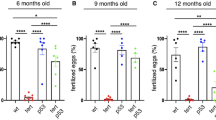
Telomere dysfunction limits the proliferative capacity of human cells and induces organismal aging by activation of p53 and p21 (refs. 1, 2, 3, 4, 5, 6). Although deletion of p21 elongates the lifespan of telomere-dysfunctional mice2, a direct analysis of p53 in telomere-related aging has been hampered by early tumor formation in p53 knockout mice6. Here we analyzed the functional consequences of conditional p53 deletion7. Intestinal deletion of p53 shortened the lifespan of telomere-dysfunctional mice without inducing tumor formation. In contrast to p21 deletion, the deletion of p53 impaired the depletion of chromosomal-instable intestinal stem cells in aging telomere-dysfunctional mice. These instable stem cells contributed to epithelial regeneration leading to an accumulation of chromosomal instability, increased apoptosis, altered epithelial cell differentiation and premature intestinal failure. Together, these results provide the first experimental evidence for an organ system in which p53-dependent mechanisms prevent tissue destruction in response to telomere dysfunction by depleting genetically instable stem cells.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
References
Wright, W.E. & Shay, J.W. The two-stage mechanism controlling cellular senescence and immortalization. Exp. Gerontol. 27, 383–389 (1992).
Choudhury, A.R. et al. Cdkn1a deletion improves stem cell function and lifespan of mice with dysfunctional telomeres without accelerating cancer formation. Nat. Genet. 39, 99–105 (2007).
Rudolph, K.L. et al. Longevity, stress response, and cancer in aging telomerase-deficient mice. Cell 96, 701–712 (1999).
Schaetzlein, S. et al. Exonuclease-1 deletion impairs DNA damage signaling and prolongs lifespan of telomere-dysfunctional mice. Cell 130, 863–877 (2007).
Hemann, M.T. et al. The shortest telomere, not average telomere length, is critical for cell viability and chromosome stability. Cell 107, 67–77 (2001).
Chin, L. et al. p53 deficiency rescues the adverse effects of telomere loss and cooperates with telomere dysfunction to accelerate carcinogenesis. Cell 97, 527–538 (1999).
Jonkers, J. et al. Synergistic tumor suppressor activity of BRCA2 and p53 in a conditional mouse model for breast cancer. Nat. Genet. 29, 418–425 (2001).
Tyner, S.D. et al. p53 mutant mice that display early ageing-associated phenotypes. Nature 415, 45–53 (2002).
Garcíía-Cao, I. et al. “Super p53” mice exhibit enhanced DNA damage response, are tumor resistant and age normally. EMBO J. 21, 6225–6235 (2002).
Matheu, A. et al. Delayed ageing through damage protection by the Arf/p53 pathway. Nature 448, 375–379 (2007).
Tomás-Loba, A. et al. Telomerase reverse transcriptase delays aging in cancer-resistant mice. Cell 135, 609–622 (2008).
Jiang, H. et al. Proteins induced by telomere dysfunction and DNA damage represent biomarkers of human aging and disease. Proc. Natl. Acad. Sci. USA 105, 11299–11304 (2008).
Lieber, M.R. & Karanjawala, Z.E. Ageing, repetitive genomes and DNA damage. Nat. Rev. Mol. Cell Biol. 5, 69–75 (2004).
Nalapareddy, K. et al. Determining the influence of telomere dysfunction and DNA damage on stem and progenitor cell aging - what markers can we use? Exp. Gerontol. 43, 998–1004 (2008).
Brown, J.P., Wei, W. & Sedivy, J.M. Bypass of senescence after disruption of p21CIP1/WAF1 gene in normal diploid human fibroblasts. Science 277, 831–834 (1997).
García-Cao, I. et al. Increased p53 activity does not accelerate telomere-driven ageing. EMBO Rep. 7, 546–552 (2006).
el Marjou, F. et al. Tissue-specific and inducible Cre-mediated recombination in the gut epithelium. Genesis 39, 186–193 (2004).
Rudolph, K.L. et al. Telomere dysfunction and evolution of intestinal carcinoma in mice and humans. Nat. Genet. 28, 155–159 (2001).
Barker, N. et al. Identification of stem cells in small intestine and colon by marker gene Lgr5. Nature 449, 1003–1007 (2007).
van der Flier, L.G. et al. Transcription factor achaete scute-like 2 controls intestinal stem cell fat. Cell 136, 903–912 (2009).
Rando, T.A. Stem cells, ageing and the quest for immortality. Nature 441, 1080–1086 (2006).
Sharpless, N.E. & DePinho, R.A. How stem cells age and why this makes us grow old. Nat. Rev. Mol. Cell Biol. 8, 703–713 (2007).
Chambers, S.M. et al. Aging hematopoietic stem cells decline in function and exhibit epigenetic dysregulation. PLoS Biol. 5, e201 (2007).
Riboni, R. et al. Telomeric fusions in cultured human fibroblasts as a source of genomic instability. Cancer Genet. Cytogenet. 95, 130–136 (1997).
Feng, Z. et al. Declining p53 function in the aging process: a possible mechanism for the increased tumor incidence in older populations. Proc. Natl. Acad. Sci. USA 104, 16633–16638 (2007).
Komarova, E.A. et al. Dual effect of p53 on radiation sensitivity in vivo: p53 promotes hematopoietic injury, but protects from gastro-intestinal syndrome in mice. Oncogene 23, 3265–3271 (2004).
Flores, I. & Blasco, M.A. A p53-dependent response limits epidermal stem cell functionality and organismal size in mice with short telomeres. PLoS One 4, e4934 (2009).
Poon, S.S. et al. Telomere length measurements using digital fluorescence microscopy. Cytometry 36, 267–278 (1999).
Chung, Y.J. et al. A whole-genome mouse BAC microarray with 1-Mb resolution for analysis of DNA copy number changes by array comparative genomic hybridization. Genome Res. 14, 188–196 (2004).
Geigl, J.B. & Speicher, M.R. Single-cell isolation from cell suspensions and whole genome amplification from single cells to provide templates for CGH analysis. Nat Protoc. 2, 3173–3184 (2007).
van Beers, E.H. et al. A multiplex PCR predictor for aCGH success of FFPE samples. Br. J. Cancer 94, 333–337 (2006).
Acknowledgements
We thank S. Robine (Institute Curie-CNRS, Paris) for providing villin-Cre-ERT2 mice and A. Berns (The Netherlands Cancer Institute, Amsterdam) for providing conditional p53 knockout mice. We thank J. Jonkers ((The Netherlands Cancer Institute, Amsterdam ) for providing the BAC clones for chromosome FISH. We thank G. Schütz (German Cancer Research Center, Heidelberg) for providing self-made Cre antibody. We thank C. Günes for critical reading of the manuscript. This project was supported by funding from the Deutsche Forschungsgemeinschaft (KFO 167, RU745/10-1), by the Deutsche Krebshilfe e.V. (consortium grant on tumor stem cells) and by the European Union (GENINCA, Telomarker). K.L.R. and M.R.S. are both supported by the European Union (GENINCA, contract number 202230).
Author information
Authors and Affiliations
Contributions
Y.B.-N. conducted experiments and data analysis; A.L. and E.P. conducted the chromosome FISH; A.C.O., M.R.S. and E.H. conducted array-CGH analysis; K.N. and P.S. did the analysis of intestinal atrophy and pathology; F.S. and B.L. conducted the laser microdissection of intestinal crypts; H.K. was responsible for the bioinformatics; E.D., N.B. and H.C. conducted the OLFM4 in situ; K.L.R. was responsible for the study design and manuscript preparation.
Corresponding author
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–6 and Supplementary Tables 1 and 2 (PDF 1143 kb)
Rights and permissions
About this article
Cite this article
Begus-Nahrmann, Y., Lechel, A., Obenauf, A. et al. p53 deletion impairs clearance of chromosomal-instable stem cells in aging telomere-dysfunctional mice. Nat Genet 41, 1138–1143 (2009). https://doi.org/10.1038/ng.426
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ng.426
This article is cited by
-
Functions and regulatory mechanisms of resting hematopoietic stem cells: a promising targeted therapeutic strategy
Stem Cell Research & Therapy (2023)
-
NRF2 signaling pathway and telomere length in aging and age-related diseases
Molecular and Cellular Biochemistry (2023)
-
Aneuploidy-inducing gene knockdowns overlap with cancer mutations and identify Orp3 as a B-cell lymphoma suppressor
Oncogene (2020)
-
Mitophagy-driven metabolic switch reprograms stem cell fate
Cellular and Molecular Life Sciences (2019)
-
Relevance of the p53–MDM2 axis to aging
Cell Death & Differentiation (2018)