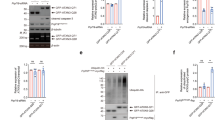
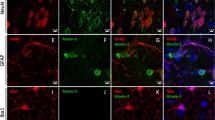
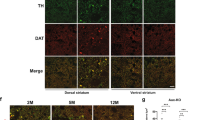
Abstract
Spinocerebellar ataxia type 1 (SCA1) is a dominantly inherited neurodegenerative disease caused by expansion of a glutamine tract in ataxin-1 (ATXN1). SCA1 pathogenesis studies support a model in which the expanded glutamine tract causes toxicity by modulating the normal activities of ATXN1. To explore native interactions that modify the toxicity of ATXN1, we generated a targeted duplication of the mouse ataxin-1-like (Atxn1l, also known as Boat) locus, a highly conserved paralog of SCA1, and tested the role of this protein in SCA1 pathology. Using a knock-in mouse model of SCA1 that recapitulates the selective neurodegeneration seen in affected individuals, we found that elevated Atxn1l levels suppress neuropathology by displacing mutant Atxn1 from its native complex with Capicua (CIC). Our results provide genetic evidence that the selective neuropathology of SCA1 arises from modulation of a core functional activity of ATXN1, and they underscore the importance of studying the paralogs of genes mutated in neurodegenerative diseases to gain insight into mechanisms of pathogenesis.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
References
Tsuda, H. et al. The AXH domain of Ataxin-1 mediates neurodegeneration through its interaction with Gfi-1/Senseless proteins. Cell 122, 633–644 (2005).
Tsai, C.C. et al. Ataxin 1, a SCA1 neurodegenerative disorder protein, is functionally linked to the silencing mediator of retinoid and thyroid hormone receptors. Proc. Natl. Acad. Sci. USA 101, 4047–4052 (2004).
Mizutani, A. et al. Boat, an AXH domain protein, suppresses the cytotoxicity of mutant ataxin-1. EMBO J. 24, 3339–3351 (2005).
Lam, Y.C. et al. ATAXIN-1 interacts with the repressor Capicua in its native complex to cause SCA1 neuropathology. Cell 127, 1335–1347 (2006).
Matilla, A. et al. Mice lacking ataxin-1 display learning deficits and decreased hippocampal paired-pulse facilitation. J. Neurosci. 18, 5508–5516 (1998).
Lim, J. et al. A protein-protein interaction network for human inherited ataxias and disorders of Purkinje cell degeneration. Cell 125, 801–814 (2006).
Watase, K. et al. A long CAG repeat in the mouse Sca1 locus replicates SCA1 features and reveals the impact of protein solubility on selective neurodegeneration. Neuron 34, 905–919 (2002).
Arrasate, M., Mitra, S., Schweitzer, E.S., Segal, M.R. & Finkbeiner, S. Inclusion body formation reduces levels of mutant huntingtin and the risk of neuronal death. Nature 431, 805–810 (2004).
Bowman, A.B., Yoo, S.Y., Dantuma, N.P. & Zoghbi, H.Y. Neuronal dysfunction in a polyglutamine disease model occurs in the absence of ubiquitin-proteasome system impairment and inversely correlates with the degree of nuclear inclusion formation. Hum. Mol. Genet. 14, 679–691 (2005).
Slow, E.J. et al. Absence of behavioral abnormalities and neurodegeneration in vivo despite widespread neuronal huntingtin inclusions. Proc. Natl. Acad. Sci. USA 102, 11402–11407 (2005).
Zhang, X. et al. A potent small molecule inhibits polyglutamine aggregation in Huntington's disease neurons and suppresses neurodegeneration in vivo. Proc. Natl. Acad. Sci. USA 102, 892–897 (2005).
Bodner, R.A. et al. Pharmacological promotion of inclusion formation: a therapeutic approach for Huntington's and Parkinson's diseases. Proc. Natl. Acad. Sci. USA 103, 4246–4251 (2006).
Taylor, J.P. et al. Aggresomes protect cells by enhancing the degradation of toxic polyglutamine-containing protein. Hum. Mol. Genet. 12, 749–757 (2003).
Klement, I.A. et al. Ataxin-1 nuclear localization and aggregation: role in polyglutamine-induced disease in SCA1 transgenic mice. Cell 95, 41–53 (1998).
Cattaneo, E., Zuccato, C. & Tartari, M. Normal huntingtin function: an alternative approach to Huntington's disease. Nat. Rev. Neurosci. 6, 919–930 (2005).
Cui, L. et al. Transcriptional repression of PGC-1alpha by mutant huntingtin leads to mitochondrial dysfunction and neurodegeneration. Cell 127, 59–69 (2006).
Zhai, W., Jeong, H., Cui, L., Krainc, D. & Tjian, R. In vitro analysis of huntingtin-mediated transcriptional repression reveals multiple transcription factor targets. Cell 123, 1241–1253 (2005).
Dragatsis, I., Levine, M.S. & Zeitlin, S. Inactivation of Hdh in the brain and testis results in progressive neurodegeneration and sterility in mice. Nat. Genet. 26, 300–306 (2000).
Van Raamsdonk, J.M. et al. Loss of wild-type huntingtin influences motor dysfunction and survival in the YAC128 mouse model of Huntington disease. Hum. Mol. Genet. 14, 1379–1392 (2005).
Auerbach, W. et al. The HD mutation causes progressive lethal neurological disease in mice expressing reduced levels of huntingtin. Hum. Mol. Genet. 10, 2515–2523 (2001).
Leavitt, B.R. et al. Wild-type huntingtin reduces the cellular toxicity of mutant huntingtin in vivo. Am. J. Hum. Genet. 68, 313–324 (2001).
Zuccato, C. et al. Huntingtin interacts with REST/NRSF to modulate the transcription of NRSE-controlled neuronal genes. Nat. Genet. 35, 76–83 (2003).
Ho, L.W., Brown, R., Maxwell, M., Wyttenbach, A. & Rubinsztein, D.C. Wild type Huntingtin reduces the cellular toxicity of mutant Huntingtin in mammalian cell models of Huntington's disease. J. Med. Genet. 38, 450–452 (2001).
Graham, R.K. et al. Cleavage at the caspase-6 site is required for neuronal dysfunction and degeneration due to mutant huntingtin. Cell 125, 1179–1191 (2006).
Gauthier, L.R. et al. Huntingtin controls neurotrophic support and survival of neurons by enhancing BDNF vesicular transport along microtubules. Cell 118, 127–138 (2004).
da Costa, C.A., Masliah, E. & Checler, F. Beta-synuclein displays an antiapoptotic p53-dependent phenotype and protects neurons from 6-hydroxydopamine-induced caspase 3 activation: cross-talk with alpha-synuclein and implication for Parkinson's disease. J. Biol. Chem. 278, 37330–37335 (2003).
Yoo, S.Y. et al. SCA7 knockin mice model human SCA7 and reveal gradual accumulation of mutant ataxin-7 in neurons and abnormalities in short-term plasticity. Neuron 37, 383–401 (2003).
Chen, H.K. et al. Interaction of Akt-phosphorylated ataxin-1 with 14–3-3 mediates neurodegeneration in spinocerebellar ataxia type 1. Cell 113, 457–468 (2003).
Copps, K. et al. Complex formation by the Drosophila MSL proteins: role of the MSL2 RING finger in protein complex assembly. EMBO J. 17, 5409–5417 (1998).
Acknowledgements
We are grateful to C.-C. Tsai (University of Medicine and Dentistry of New Jersey—Robert Wood Johnson Medical School) for providing ATXN1L antiserum; S. Vaishnav for mouse genotyping; A. McCall for isolating and characterizing Atxn1l clones; G. Schuster for ES cell work and generation of chimeric animals; K. Schulze for artwork (Fig. 6) and A. Flora, J. Lim, J. Gatchel, P. Moretti, E. Hyun and J. Crespo-Barreto for many discussions. This work was supported by the following US National Institutes of Health grants: National Institute for Neurological Disorders and Stroke grants NS27699 (to H.Y.Z.), NS22920 and NS45667 (to H.T.O.) and National Institute of Child Health and Human Development grant HD024064 (to the Baylor College of Medicine Mental Retardation and Developmental Disabilities Research Center). A.B.B. was a postdoctoral fellow of the Hereditary Disease Foundation. H.Y.Z. is a Howard Hughes Medical Institute investigator.
Author information
Authors and Affiliations
Contributions
A.B. designed, performed or assisted in most of the experiments; analyzed data; characterized the Atxn1l duplication allele and wrote the manuscript. Y.L. performed ATXN1L immunoprecipitation experiments, identified the CIC protein interaction, designed and characterized the CIC antisera and edited the manuscript. P.J. designed and performed cerebellar Atxn1 immunoprecipitation experiments, characterized Atxn1l sera and provided critical commentary. H.C. designed and constructed the targeting construct. R.R. performed cerebellar extraction, protein blots and HPLC chromatography and characterized Atxn1l sera. R.S. performed the in situ hybridization experiments. J.F. performed cerebellar CIC immunoprecipitation experiments, provided critical commentary and edited the manuscript. J.K. made the ATXN1L cDNA expression vector. H.O. assisted in data analysis and interpretation. H.Z. led the project by inspiring the search for ATXN1 paralogs, generated major hypotheses and formulated the genetic and biochemical experiments, interpreted data, provided experimental direction and edited the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Fig. 1
Atxn1l expression in Purkinje cells of adult mouse cerebellum. (PDF 527 kb)
Supplementary Fig. 2
Fractionation of ATXN1L, ATXN1 and CIC by gel filtration chromatography of mouse cerebellar extracts. (PDF 176 kb)
Supplementary Fig. 3
ATXN1 and ATXN1L compete for interaction with CIC. (PDF 106 kb)
Supplementary Fig. 4
Atxn1l duplication suppresses Sca1154Q/+ cerebellar pathology. (PDF 735 kb)
Supplementary Fig. 5
Atxn1l duplication suppresses Sca1154Q/+ lethality. (PDF 341 kb)
Supplementary Fig. 6
ATXN1L levels are similar between wild-type and Sca1154Q/+ animals. (PDF 131 kb)
Supplementary Table 1
ANOVA post hoc pairwise comparisons. (PDF 55 kb)
Rights and permissions
About this article
Cite this article
Bowman, A., Lam, Y., Jafar-Nejad, P. et al. Duplication of Atxn1l suppresses SCA1 neuropathology by decreasing incorporation of polyglutamine-expanded ataxin-1 into native complexes. Nat Genet 39, 373–379 (2007). https://doi.org/10.1038/ng1977
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ng1977
This article is cited by
-
Functional implications of paralog genes in polyglutamine spinocerebellar ataxias
Human Genetics (2023)
-
Regulation and function of capicua in mammals
Experimental & Molecular Medicine (2020)
-
Pathogenic mechanisms underlying spinocerebellar ataxia type 1
Cellular and Molecular Life Sciences (2020)
-
Transcriptomic analysis of CIC and ATXN1L reveal a functional relationship exploited by cancer
Oncogene (2019)
-
Molecular Mechanisms and Therapeutics for SCA17
Neurotherapeutics (2019)