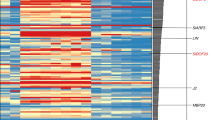
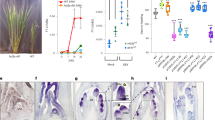
Abstract
In maize (Zea mays), sex determination occurs through abortion of female carpels in the tassel and arrest of male stamens in the ear. The Tasselseed6 (Ts6) and tasselseed4 (ts4) mutations permit carpel development in the tassel while increasing meristem branching, showing that sex determination and acquisition of meristem fate share a common pathway. We show that ts4 encodes a mir172 microRNA that targets APETALA2 floral homeotic transcription factors. Three lines of evidence suggest that indeterminate spikelet1 (ids1), an APETALA2 gene required for spikelet meristem determinacy, is a key target of ts4. First, loss of ids1 suppresses the ts4 sex determination and branching defects. Second, Ts6 mutants phenocopy ts4 and possess mutations in the microRNA binding site of ids1. Finally, IDS1 protein is expressed more broadly in ts4 mutants compared to wild type. Our results demonstrate that sexual identity in maize is acquired by limiting floral growth through negative regulation of the floral homeotic pathway.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
References
Phipps, I.F. Heritable characters in maize. XXXI. Tassel-seed4. J. Hered. 19, 399–404 (1928).
Veit, B., Schmidt, R.J., Hake, S. & Yanofsky, M.F. Maize floral development–new genes and old mutants. Plant Cell 5, 1205–1215 (1993).
DeLong, A., Calderon-Urrea, A. & Dellaporta, S.L. Sex determination gene TASSELSEED2 of maize encodes a short-chain alcohol dehydrogenase required for stage-specific floral organ abortion. Cell 74, 757–768 (1993).
Irish, E.E., Langdale, J.A. & Nelson, T.M. Interactions between tasselseed genes and other sex determining genes in maize. Dev. Genet. 15, 155–171 (1994).
Cheng, P.C., Greyson, R.I. & Walden, D.B. Organ initiation and the development of unisexual flowers in the tassel and ear of Zea mays. Am. J. Bot. 70, 450–462 (1983).
Bortiri, E. et al. ramosa2 encodes a Lateral Organ Boundary domain protein that determines the fate of stem cells in branch meristems of maize. Plant Cell 18, 574–585 (2006).
Moore, G., Devos, K.M., Wang, Z. & Gale, M.D. Cereal genome evolution - grasses, line up and form a circle. Curr. Biol. 7, 737–739 (1995).
Irish, E.E. Class II tassel seed mutations provide evidence for multiple types of inflorescence meristems in maize (Poaceae). Am. J. Bot. 84, 1502–1515 (1997).
Xie, Z. et al. Expression of Arabidopsis MIRNA genes. Plant Physiol. 138, 2145–2154 (2005).
Reeves, R. & Wolffe, A.P. Substrate structure influences binding of the non-histone protein HMG-I(Y) to free nucleosomal DNA. Biochemistry 35, 5063–5074 (1996).
Bensen, R.J. et al. Cloning and characterization of the maize An1 gene. Plant Cell 7, 75–84 (1995).
Lauter, N., Kampani, A., Carlson, S., Goebel, M. & Moose, S.P. microRNA172 down-regulates glossy15 to promote vegetative phase change in maize. Proc. Natl. Acad. Sci. USA 102, 9412–9417 (2005).
Chuck, G., Meeley, R. & Hake, S. The control of maize spikelet meristem fate by the APETALA2-like gene indeterminate spikelet1. Genes Dev. 12, 1145–1154 (1998).
Simons, K.J. et al. Molecular characterization of the major wheat domestication gene Q. Genetics 172, 547–555 (2006).
Schwab, R., Palatnik, J., Riester, M., Schommer, C., Schmid, M. & Weigel, D. Specific effects of microRNAs on the plant transcriptome. Dev. Cell 8, 517–527 (2005).
Chen, X. A microRNA as a translational repressor of APETALA2 in Arabidopsis flower development. Science 303, 2022–2025 (2004).
Aukerman, M.J. & Sakai, H. Regulation of flowering time and floral organ identity by a microRNA and its APETALA2-like target genes. Plant Cell 15, 2730–2741 (2003).
Mizukami, Y. & Ma, H. Ectopic expression of the floral homeotic gene AGAMOUS in transgenic Arabidopsis plants alters floral organ identity. Cell 71, 119–131 (1992).
Zhao, L., Kim, Y., Dinh, T.T. & Chen, X. miR172 regulates stem cell fate and defines the inner boundary of APETALA3 and PISTILLATA expression domain in Arabidopsis floral meristems. Plant J. 51, 840–849 (2007).
Nickerson, N.H. Sustained treatment with gibberellin acid of five different kinds of maize. Ann. Mo. Bot. Gard. 46, 19–37 (1959).
Okamuro, J.K., Szeto, W., Lotys-Prass, C. & Jofuku, K.D. Photo and hormonal control of meristem identity in the Arabidopsis flower mutants apetala2 and apetala1. Plant Cell 9, 37–47 (1997).
Postlethwait, S.N. & Nelson, O.E. Characterization of development in maize through the use of mutants I. The polytypic (Pt) and ramosa-1 (ra1) mutants. Am. J. Bot. 51, 238–243 (1964).
Park, W., Li, J., Song, R., Messing, J. & Chen, X. CARPEL FACTORY, a Dicer homolog, and HEN1, a novel protein, act in microRNA metabolism in Arabidopsis thaliana. Curr. Biol. 12, 1484–1495 (2002).
Jackson, D. In situ hybridization in plants. in Molecular Plant Pathology: a Practical Approach (eds. Bowles, D.J., Gurr, S.J. & McPherson, M.) 163–174 (Oxford Univ. Press, Oxford, 1991).
Kaplinsky, N.J. & Freeling, M. Combinatorial control of meristem identity in maize inflorescences. Development 130, 1149–1158 (2003).
Jackson, D., Veit, B. & Hake, S. Expression of maize KNOTTED1 related homeobox genes in the shoot apical meristem predicts patterns of morphogenesis in the vegetative shoot. Development 120, 405–413 (1994).
Acknowledgements
The work was supported by National Science Foundation (NSF) grant DBI-0604923, by USDA-ARS Current Research Information System (CRIS) grant 5335-21000-018-00D to S.H. and by Cooperative State Research, Education, and Extension Service (CSREES) grant 2004-35301-14507 to G.C. We thank D. Irvine for the use of the scanning electron microscopy facility; D. Hantz for greenhouse maintenance; T. Peterson for the gift of the ts4-TP allele; M. Sachs for determining the genealogy of ts4-ref; B. Thompson, C. Lunde and E. Bortiri for helpful comments on the manuscript and L. Bartling and K. Saeturn for technical assistance.
Author information
Authors and Affiliations
Contributions
R.M. contributed the ts4-mum1 allele, E.I. contributed the ts4-TP and ts4-A alleles and H.S. carried out BAC sequencing. G.C. designed this study and wrote the manuscript with help from S.H.
Corresponding author
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–3, Supplementary Table 1 (PDF 1522 kb)
Rights and permissions
About this article
Cite this article
Chuck, G., Meeley, R., Irish, E. et al. The maize tasselseed4 microRNA controls sex determination and meristem cell fate by targeting Tasselseed6/indeterminate spikelet1. Nat Genet 39, 1517–1521 (2007). https://doi.org/10.1038/ng.2007.20
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ng.2007.20
This article is cited by
-
Identification and analysis of miRNAs differentially expressed in male and female Trichosanthes kirilowii maxim
BMC Genomics (2023)
-
Implications of small RNAs in plant development, abiotic stress response and crop improvement in changing climate
The Nucleus (2023)
-
Novel insights into maize (Zea mays) development and organogenesis for agricultural optimization
Planta (2023)
-
Factors specifying sex determination in maize
Plant Reproduction (2023)
-
Dosage-sensitive miRNAs trigger modulation of gene expression during genomic imbalance in maize
Nature Communications (2022)