Abstract
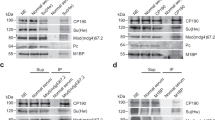
RNA interference (RNAi) is a conserved silencing mechanism that can act through alteration of chromatin structure. Chromatin insulators promote higher-order nuclear organization, thereby establishing DNA domains subject to distinct transcriptional controls. We present evidence for a functional relationship between RNAi and the gypsy insulator of D. melanogaster. Insulator activity is decreased when Argonaute genes required for RNAi are mutated, and insulator function is improved when the levels of the Rm62 helicase, involved in double-stranded RNA (dsRNA)-mediated silencing and heterochromatin formation, are reduced. Rm62 interacts physically with the DNA-binding insulator protein CP190 in an RNA-dependent manner. Finally, reduction of Rm62 levels results in marked nuclear reorganization of a compromised insulator. These results suggest that the RNAi machinery acts as a modulator of nuclear architecture capable of effecting global changes in gene expression.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
References
Volpe, T.A. et al. Regulation of heterochromatic silencing and histone H3 lysine-9 methylation by RNAi. Science 297, 1833–1837 (2002).
Pal-Bhadra, M. et al. Heterochromatic silencing and HP1 localization in Drosophila are dependent on the RNAi machinery. Science 303, 669–672 (2004).
Hall, I.M., Noma, K. & Grewal, S.I. RNA interference machinery regulates chromosome dynamics during mitosis and meiosis in fission yeast. Proc. Natl. Acad. Sci. USA 100, 193–198 (2003).
Grimaud, C. et al. RNAi components are required for nuclear clustering of Polycomb group response elements. Cell 124, 957–971 (2006).
Parkhurst, S.M. et al. The Drosophila su(Hw) gene, which controls the phenotypic effect of the gypsy transposable element, encodes a putative DNA-binding protein. Genes Dev. 2, 1205–1215 (1988).
Pai, C.Y., Lei, E.P., Ghosh, D. & Corces, V.G. The centrosomal protein CP190 is a component of the gypsy chromatin insulator. Mol. Cell 16, 737–748 (2004).
Gerasimova, T.I., Gdula, D.A., Gerasimov, D.V., Simonova, O. & Corces, V.G. A Drosophila protein that imparts directionality on a chromatin insulator is an enhancer of position-effect variegation. Cell 82, 587–597 (1995).
Gerasimova, T.I., Byrd, K. & Corces, V.G. A chromatin insulator determines the nuclear localization of DNA. Mol. Cell 6, 1025–1035 (2000).
Capelson, M. & Corces, V.G. The ubiquitin ligase dTopors directs the nuclear organization of a chromatin insulator. Mol. Cell 20, 105–116 (2005).
Byrd, K. & Corces, V.G. Visualization of chromatin domains created by the gypsy insulator of Drosophila. J. Cell Biol. 162, 565–574 (2003).
Yusufzai, T.M. & Felsenfeld, G. The 5′-HS4 chicken beta-globin insulator is a CTCF-dependent nuclear matrix-associated element. Proc. Natl. Acad. Sci. USA 101, 8620–8624 (2004).
Ishizuka, A., Siomi, M.C. & Siomi, H. A Drosophila fragile X protein interacts with components of RNAi and ribosomal proteins. Genes Dev. 16, 2497–2508 (2002).
Csink, A.K., Linsk, R. & Birchler, J.A. The Lighten up (Lip) gene of Drosophila melanogaster, a modifier of retroelement expression, position effect variegation and white locus insertion alleles. Genetics 138, 153–163 (1994).
Parkhurst, S.M. & Corces, V.G. Interactions among the gypsy transposable element and the yellow and the suppressor of hairy-wing loci in Drosophila melanogaster. Mol. Cell. Biol. 6, 47–53 (1986).
Jack, J.W. Molecular organization of the cut locus of Drosophila melanogaster. Cell 42, 869–876 (1985).
Tsai, S.F. et al. Gypsy retrotransposon as a tool for the in vivo analysis of the regulatory region of the optomotor-blind gene in Drosophila. Proc. Natl. Acad. Sci. USA 94, 3837–3841 (1997).
Schupbach, T. & Wieschaus, E. Female sterile mutations on the second chromosome of Drosophila melanogaster. II. Mutations blocking oogenesis or altering egg morphology. Genetics 129, 1119–1136 (1991).
Lin, H. & Spradling, A.C. A novel group of pumilio mutations affects the asymmetric division of germline stem cells in the Drosophila ovary. Development 124, 2463–2476 (1997).
Forstemann, K. et al. Normal microRNA maturation and germ-line stem cell maintenance requires Loquacious, a double-stranded RNA-binding domain protein. PLoS Biol. 3, e236 (2005).
Saito, K., Ishizuka, A., Siomi, H. & Siomi, M.C. Processing of pre-microRNAs by the Dicer-1-Loquacious complex in Drosophila cells. PLoS Biol. 3, e235 (2005).
Noma, K. et al. RITS acts in cis to promote RNA interference-mediated transcriptional and post-transcriptional silencing. Nat. Genet. 36, 1174–1180 (2004).
Cox, D.N., Chao, A. & Lin, H. piwi encodes a nucleoplasmic factor whose activity modulates the number and division rate of germline stem cells. Development 127, 503–514 (2000).
Pal-Bhadra, M., Bhadra, U. & Birchler, J.A. RNAi related mechanisms affect both transcriptional and posttranscriptional transgene silencing in Drosophila. Mol. Cell 9, 315–327 (2002).
Buszczak, M. & Spradling, A.C. The Drosophila P68 RNA helicase regulates transcriptional deactivation by promoting RNA release from chromatin. Genes Dev. 20, 977–989 (2006).
Fey, E.G., Krochmalnic, G. & Penman, S. The nonchromatin substructures of the nucleus: the ribonucleoprotein (RNP)-containing and RNP-depleted matrices analyzed by sequential fractionation and resinless section electron microscopy. J. Cell Biol. 102, 1654–1665 (1986).
Shaffer, C.D., Wuller, J.M. & Elgin, S.C.R. Methods in Cell Biology (Academic, San Diego, 1994).
Harlow, E. & Lane, D. Antibodies: A Laboratory Manual (Cold Spring Harbor Laboratory, New York, 1988).
Mongelard, F., Labrador, M., Baxter, E.M., Gerasimova, T.I. & Corces, V.G. Trans-splicing as a novel mechanism to explain interallelic complementation in Drosophila. Genetics 160, 1481–1487 (2002).
Mal'ceva, N.I., Belyaeva, E.S., King, R.C. & Zhimulev, I.F. Nurse cell polytene chromosomes of Drosophila melanogaster otu mutants: morphological changes accompanying interallelic complementation and position effect variegation. Dev. Genet. 20, 163–174 (1997).
Amero, S.A., Elgin, S.C. & Beyer, A.L. A unique zinc finger protein is associated preferentially with active ecdysone-responsive loci in Drosophila. Genes Dev. 5, 188–200 (1991).
Acknowledgements
We would like to thank A. Beyer for α-Pep and F. Fuller-Pace for α-p68; J. Birchler, S. Hou, H. Lin, and P. Macdonald for strains and Y. Zheng for fly cages. We are indebted to E. Baxter for assistance with population cages; M. Capelson and members of the Corces laboratory for discussions and J. Birchler, M. Capelson, and C. Karam for comments on the manuscript. E.P.L. is a fellow of The Jane Coffin Childs Memorial Fund for Medical Research. This work was supported by grants from the US National Institutes of Health to V.G.C.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Fig. 1
Localization of insulator proteins to a site of gypsy insertion in RNAi mutants. (PDF 217 kb)
Supplementary Fig. 2
Nuclear localization of Rm62 in diploid cells. (PDF 215 kb)
Rights and permissions
About this article
Cite this article
Lei, E., Corces, V. RNA interference machinery influences the nuclear organization of a chromatin insulator. Nat Genet 38, 936–941 (2006). https://doi.org/10.1038/ng1850
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ng1850
This article is cited by
-
A cohesin traffic pattern genetically linked to gene regulation
Nature Structural & Molecular Biology (2022)
-
M1BP cooperates with CP190 to activate transcription at TAD borders and promote chromatin insulator activity
Nature Communications (2021)
-
EAST affects the activity of Su(Hw) insulators by two different mechanisms in Drosophila melanogaster
Chromosoma (2017)
-
Heterochromatin-binding proteins regulate male X polytene chromosome morphology and dosage compensation: an evidence from a variegated rearranged strain [In (1)BM 2,(rv)] and its interactions with hyperploids and mle mutation in Drosophila melanogaster
The Nucleus (2016)
-
Morphology of nuclear transcription
Histochemistry and Cell Biology (2016)